
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,797,983 – 19,798,090 |
| Length | 107 |
| Max. P | 0.836517 |

| Location | 19,797,983 – 19,798,090 |
|---|---|
| Length | 107 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.97 |
| Shannon entropy | 0.53240 |
| G+C content | 0.44328 |
| Mean single sequence MFE | -24.68 |
| Consensus MFE | -13.78 |
| Energy contribution | -13.75 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.836517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
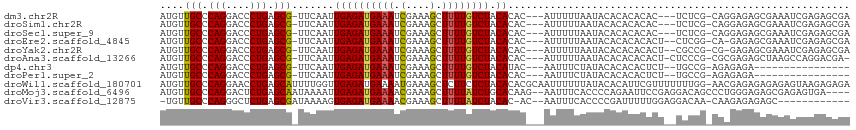
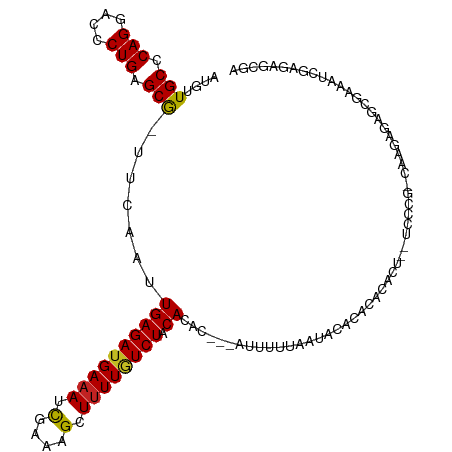
>dm3.chr2R 19797983 107 - 21146708 AUGUUGCCCAGGACCCUGAGCG-UUCAAUUGAGAUGAAAUCGAAAGCUUUUGUCUACACAC---AUUUUUAAUACACACACAC---UCUCG-CAGGAGAGCGAAAUCGAGAGCGA .....((.(((....))).))(-(((...(((((..(((.(....).)))..))).))...---................(.(---((((.-...))))).).......)))).. ( -26.30, z-score = -1.37, R) >droSim1.chr2R 18357385 107 - 19596830 AUGUUGCCCAGGACCCUGAGCG-UUCAAUUGAGAUGAAAUCGAAAGCUUUUGUCUACACAC---AUUUUUAAUACACACACAC---UCUCG-CAGGAGAGCGAAAUCGAGAGCGA .....((.(((....))).))(-(((...(((((..(((.(....).)))..))).))...---................(.(---((((.-...))))).).......)))).. ( -26.30, z-score = -1.37, R) >droSec1.super_9 3089581 107 - 3197100 AUGUUGCCCAGGACCCUGAGCG-UUCAAUUGAGAUGAAAUCGAAAGCUUUUGUCUACACAC---AUUUUUAAUACACACACAC---UCUCG-CAGGAGAGCGAAAUCGAGAGCGA .....((.(((....))).))(-(((...(((((..(((.(....).)))..))).))...---................(.(---((((.-...))))).).......)))).. ( -26.30, z-score = -1.37, R) >droEre2.scaffold_4845 21178358 107 - 22589142 AUGUUGCCCAGGACCCUGAGCG-UUCAAUUGAGAUGAAAUCGAAAGCUUUUGUCUACACAC---AUUUUUAAUACACACACACU--CUCGG-CA-GAGAGCGAAAUCGAGAGCGA .....((.(((....))).))(-(((...(((((..(((.(....).)))..))).))...---................(.((--(((..-..-))))).).......)))).. ( -26.90, z-score = -1.57, R) >droYak2.chr2R 19783124 107 - 21139217 AUGUUGCCCAGGACCCUGAGCG-UUCAAUUGAGAUGAAAUCGAAAGCUUUUGUCUACACAC---AUUUUUAAUACACACACACU--CGCCG-CG-GAGAGCGAAAUCGAGAGCGA .....((.(((....))).))(-(((...(((((..(((.(....).)))..))).))...---...................(--((((.-..-..).))))......)))).. ( -24.00, z-score = -0.54, R) >droAna3.scaffold_13266 5669606 108 - 19884421 AUGUUGCCCAGGACCCUGAGCG-UUCAAUUGAGAUGAAAUCGAAAGCUUUUGUCUACACAC---AUUUUUAAUACACACACACU-CUCCCG-CGCGAGAGCUAAGCCAGGACGA- .....((.(((....))).))(-(((...(((((..(((.(....).)))..))).))...---..................((-(((...-...)))))........))))..- ( -24.20, z-score = -1.08, R) >dp4.chr3 1746220 92 - 19779522 AUGUUGCCCAGGACCCUGAGCG-UUCAAUUGAGAUGAAAUCGAAAGCUUUUGUCUACAUAC---AAUUUCUAUACACACACUCU--UGCCG-AGAGAGA---------------- .((.(((.(((....))).)))-..))..(((((..(((.(....).)))..))).))...---................((((--(....-.))))).---------------- ( -19.70, z-score = -1.19, R) >droPer1.super_2 1933793 92 - 9036312 AUGUUGCCCAGGACCCUGAGCG-UUCAAUUGAGAUGAAAUCGAAAGCUUUUGUCUACACAC---AAUUUCUAUACACACACUCU--UGCCG-AGAGAGA---------------- .((.(((.(((....))).)))-..))..(((((..(((.(....).)))..))).))...---................((((--(....-.))))).---------------- ( -19.60, z-score = -1.04, R) >droWil1.scaffold_180701 2718839 114 + 3904529 AUGUUGCCCAGGAACCUGAGCAUUUUGGUUGAGAUGAAAAUGAAAGCUCUUCUCUACACACGCAAUUUUUUAUACACAUUCGUUUUUUUGG-AACGAGAGAGAGAGUAAGAGAGA ....(((.(((....))).))).(((.(((........))).)))(((((.((((........((....)).......(((((((.....)-)))))))))))))))........ ( -20.90, z-score = 0.74, R) >droMoj3.scaffold_6496 14940376 109 - 26866924 AUGUUGCCCAGGACUCUGAGCAAUAAAAUUGAGAUGAAAACGAAAGCUUUUAUCUGCACAAG--AAUUUCACCCCAGAAUUCCGAGGACAGCCCUGGGAGAGCGAGAGUGA---- .((((((.(((....))).))))))....((((((((((.(....).)))))))).))....--....((((.(..(..((((.(((.....))).))))..)..).))))---- ( -31.80, z-score = -2.79, R) >droVir3.scaffold_12875 11943241 98 - 20611582 -UGUUGCCCAGGGCUCUGAGCGAUAAAAGUGAGAUGAAAACGAAAGCUUUUAUCUACAC-AC--AAUUUCACCCCGAUUUUUGGAGGACAA-CAAGAGAGAGC------------ -((((((.(((....))).))))))...(((((((((((.(....).)))))))).)))-..--.......((((((...)))).))....-...........------------ ( -25.50, z-score = -2.34, R) >consensus AUGUUGCCCAGGACCCUGAGCG_UUCAAUUGAGAUGAAAUCGAAAGCUUUUGUCUACACAC___AUUUUUAAUACACACACACU__UCCCG_CAAGAGAGCGAAAUCGAGAGCGA .....((.(((....))).))........((((((((((.(....).)))))))).))......................................................... (-13.78 = -13.75 + -0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:46:50 2011