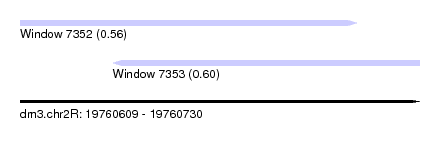
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,760,609 – 19,760,730 |
| Length | 121 |
| Max. P | 0.604420 |

| Location | 19,760,609 – 19,760,711 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 88.60 |
| Shannon entropy | 0.21535 |
| G+C content | 0.50882 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -23.38 |
| Energy contribution | -23.97 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.559435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
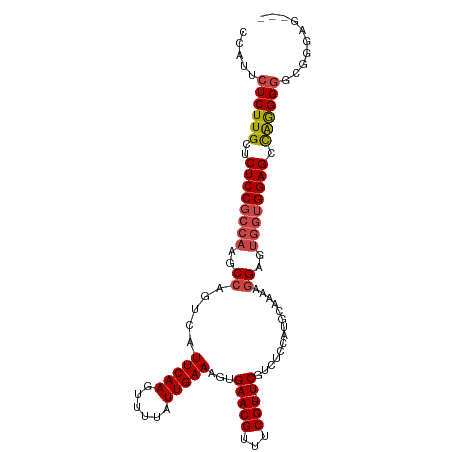
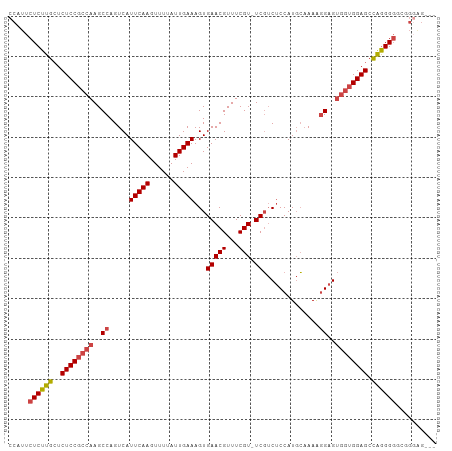
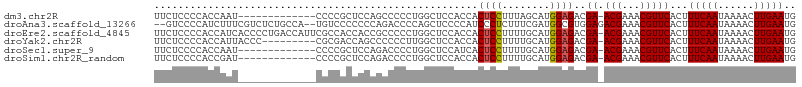
>dm3.chr2R 19760609 102 + 21146708 CCAUUCUCUUGCUCUCCGCCAAGCCAGUCAUUCAAGUUUUAUUGAAAGUGAACGUUUCGU-UCGUCUCCAUGCUAAAGGAGUGGUGGAGCCAGGGGGCUGGAG--- (((..((((((..((((((((..((.....(((((......)))))((((((((...)))-))).))..........))..)))))))).))))))..)))..--- ( -37.70, z-score = -2.96, R) >droAna3.scaffold_13266 5632230 101 + 19884421 CCAUUCUCUUGAUCUCCGCCAAGCCAGUCAUUCAAGUUUUAUUGAAAGUGAACGUUUCGUCUCCACGCCAUCGAAAGAGGGAUGGGGAGCUGGGGUCUGGG----- .....(((..((((((.((.((((...((((((((......))))..))))..))))..((((((..((.((....))))..)))))))).)))))).)))----- ( -30.00, z-score = -0.15, R) >droEre2.scaffold_4845 21140999 105 + 22589142 CCAUUCUCUUGCUCUCCGCCAAGCCAGUCAUUCAAGUUUUAUUGAAAGUGAACGUUUCGU-UCGUCUCCAUGCAAAAGGAGUGGUGGAGCCAGGGGGCGGUGGUGG (((((((((((..((((((((..((.....(((((......)))))((((((((...)))-))).))..........))..)))))))).)))))))..))))... ( -36.40, z-score = -2.03, R) >droYak2.chr2R 19745977 105 + 21139217 CCAUUCUCUUGCUCUCCGCCAAGCCAGUCAUUCAAGUUUUAUUGAAAGUGAACGUUUCGU-UCGUCUCCAUGCAAAAGGAGUGGUGGAGCCAAGGGGGGCUGGUCG (((((((((((..((((((((..((.....(((((......)))))((((((((...)))-))).))..........))..)))))))).))))))))..)))... ( -35.80, z-score = -2.23, R) >droSec1.super_9 3051873 102 + 3197100 CCAUUCUCUUGCUCUCCGCCAAGCCAGUCAUUCAAGUUUUAUUGAAAGUGAACGUUUCGU-UCGUCUCCAUGCAAAAGGAGUGAUGGAGCCAGGGGUCUGGAG--- (((..((((((..(((((.((..((.....(((((......)))))((((((((...)))-))).))..........))..)).))))).))))))..)))..--- ( -30.10, z-score = -1.39, R) >droSim1.chr2R_random 2881150 102 + 2996586 CCAUUCUCUUGCUCUCCGCCAAGCCAGUCAUUCAAGUUUUAUUGAAAGUGAACGUUUCGU-UCGUCUCCAUGCAAAAGGAGUGGUGGAGCCAGGGGUCUGGAG--- (((..((((((..((((((((..((.....(((((......)))))((((((((...)))-))).))..........))..)))))))).))))))..)))..--- ( -37.00, z-score = -3.39, R) >consensus CCAUUCUCUUGCUCUCCGCCAAGCCAGUCAUUCAAGUUUUAUUGAAAGUGAACGUUUCGU_UCGUCUCCAUGCAAAAGGAGUGGUGGAGCCAGGGGGCGGGAG___ .....((((((..(((((((((((...((((((((......))))..))))..))))........((((........)))).))))))).)))))).......... (-23.38 = -23.97 + 0.59)

| Location | 19,760,637 – 19,760,730 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.24 |
| Shannon entropy | 0.39560 |
| G+C content | 0.51744 |
| Mean single sequence MFE | -17.50 |
| Consensus MFE | -6.54 |
| Energy contribution | -6.57 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.604420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19760637 93 - 21146708 UUCUCCCCACCAAU-------------CCCCGCUCCAGCCCCCUGGCUCCACCACUCCUUUAGCAUGGAGACGA-ACGAAACGUUCACUUUCAAUAAAACUUGAAUG ..............-------------.........((((....))))......((((........))))..((-(((...)))))...(((((......))))).. ( -15.40, z-score = -2.14, R) >droAna3.scaffold_13266 5632258 103 - 19884421 --GUCCCCAUCUUUCGUCUCUGCCA--UGUCCCCCCCAGACCCCAGCUCCCCAUCCCUCUUUCGAUGGCGUGGAGACGAAACGUUCACUUUCAAUAAAACUUGAAUG --.........(((((((((..(..--.(((.......))).........(((((........))))).)..)))))))))((((((..((......))..)))))) ( -24.30, z-score = -3.57, R) >droEre2.scaffold_4845 21141027 106 - 22589142 UUCUCCCCACCAUCACCCCUGACCAUUCGCCACCACCGCCCCCUGGCUCCACCACUCCUUUUGCAUGGAGACGA-ACGAAACGUUCACUUUCAAUAAAACUUGAAUG .......................(((((((((...........)))).......((((........))))..((-(((...)))))................))))) ( -17.40, z-score = -2.86, R) >droYak2.chr2R 19746005 97 - 21139217 UUCUCCCCACCAUUACCC---------CGCGACCAGCCCCCCUUGGCUCCACCACUCCUUUUGCAUGGAGACGA-ACGAAACGUUCACUUUCAAUAAAACUUGAAUG ..................---------.......((((......))))......((((........))))..((-(((...)))))...(((((......))))).. ( -15.60, z-score = -2.19, R) >droSec1.super_9 3051901 93 - 3197100 UUCUCCCCACCAAU-------------CCCCGCUCCAGACCCCUGGCUCCAUCACUCCUUUUGCAUGGAGACGA-ACGAAACGUUCACUUUCAAUAAAACUUGAAUG ..............-------------.......((((....))))((((((((.......)).))))))..((-(((...)))))...(((((......))))).. ( -16.90, z-score = -2.55, R) >droSim1.chr2R_random 2881178 93 - 2996586 UUCUCCCCACCGAU-------------CCCCGCUCCAGACCCCUGGCUCCACCACUCCUUUUGCAUGGAGACGA-ACGAAACGUUCACUUUCAAUAAAACUUGAAUG ..............-------------.......((((....))))........((((........))))..((-(((...)))))...(((((......))))).. ( -15.40, z-score = -1.72, R) >consensus UUCUCCCCACCAAU_____________CCCCGCCCCAGCCCCCUGGCUCCACCACUCCUUUUGCAUGGAGACGA_ACGAAACGUUCACUUUCAAUAAAACUUGAAUG ......................................................((((........))))...........((((((..((......))..)))))) ( -6.54 = -6.57 + 0.03)
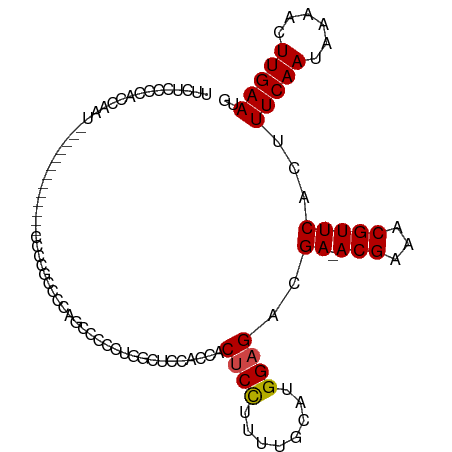
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:46:48 2011