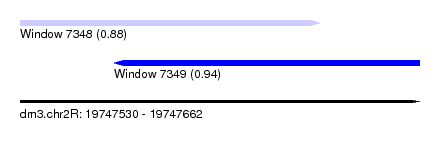
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,747,530 – 19,747,662 |
| Length | 132 |
| Max. P | 0.942909 |

| Location | 19,747,530 – 19,747,629 |
|---|---|
| Length | 99 |
| Sequences | 14 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 69.82 |
| Shannon entropy | 0.61931 |
| G+C content | 0.54752 |
| Mean single sequence MFE | -33.51 |
| Consensus MFE | -12.86 |
| Energy contribution | -12.63 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.71 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.876636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
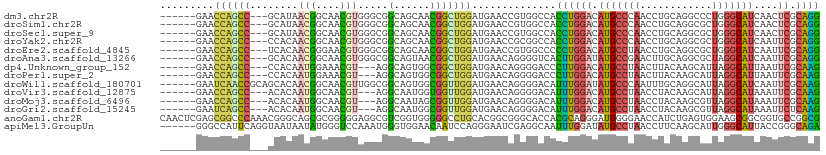
>dm3.chr2R 19747530 99 + 21146708 ---------AUGUUUUGGCCCUGGUUGGAGGAGCUCGAUCCCUGCGAGUUGAUGCCCAGGGCCUGCAGGUUGGGCAUGUCCAGGUGGCCACGGUUCAUCCAGCCGUUG--------- ---------.............(((((((.(((((.(..(((((.((....(((((((..(((....)))))))))).)))))).)..)..))))).)))))))....--------- ( -38.60, z-score = -0.05, R) >droSim1.chr2R 18311016 99 + 19596830 ---------AUGUUCUGGCCCUGGUUGGAGGAGCUCGAGCCCUGCGAGUUGAUGCCCAGCGCCUGCAGGUUGGGCAUGUCCAGGUGGCCACGGUUCAUCCAGCCGUUG--------- ---------.......(((((((((((.(((.((....))))).)))....(((((((((........)))))))))..))))..))))((((((.....))))))..--------- ( -41.50, z-score = -0.64, R) >droSec1.super_9 3039026 99 + 3197100 ---------AUGUUCUGGCCCUGGUUGGAGGAGCUCGAUCCCUGCGAGUUGAUGCCCAGCGCCUGCAGGUUGGGCAUGUCCAGGUGGCCACGGUUCAUCCAGCCGUUG--------- ---------.............(((((((.(((((.(..(((((.((....(((((((((........))))))))).)))))).)..)..))))).)))))))....--------- ( -39.60, z-score = -0.36, R) >droYak2.chr2R 19730114 99 + 21139217 ---------AUGUUCUGGCCCUGGUUGGAGGAGCUUGAUCCCUGCGAAUUGAUGCCCAGCGCCUGCAGGUUGGGCAUGUCCAGGUGGCCGCGGUUCAUCCAGCCGUUG--------- ---------.............(((((((.(((((((..(((((.((....(((((((((........))))))))).)))))).)..)).))))).)))))))....--------- ( -41.80, z-score = -1.13, R) >droEre2.scaffold_4845 21125158 99 + 22589142 ---------AUGUUCUGGCCCUGGUUUGAUGAGCUUGAUCCCUGCGAAUUGAUGCCCAGCGCCUGCAGGUUAGGCAUGUCCAGGGGGCCACGGUUCAUCCAGCCGUUG--------- ---------.............((((.((((((((((.((((((.((....(((((....(((....)))..))))).))))))))..)).)))))))).))))....--------- ( -36.40, z-score = -0.40, R) >droAna3.scaffold_13266 426412 99 + 19884421 ---------AUGUUCUGACCUUGAUUCGAUGAACUGGAACCCUGCGAAUUGAUGCCUAGCGCCUGCAAGUUCGGCAUAUCCAAGUGACCCCUGUUCAUCCAGCCGUUA--------- ---------..................(((((((.((.((..((.((....(((((.(((........))).))))).)))).))....)).))))))).........--------- ( -20.80, z-score = -0.19, R) >dp4.Unknown_group_152 9382 99 + 20572 ---------AUGUUCUGUCCUUGGUUGGACGAGCUCGAUCCUUGCGAAUUAAUGCCUAAUGCUUGUAAGUUAGGCAUGUCCAAGGGUCCCCUGUUCAUCCAGCCGCCA--------- ---------.............(((((((.((((..((((((((.((....(((((((((........))))))))).))))))))))....)))).)))))))....--------- ( -41.90, z-score = -5.82, R) >droPer1.super_2 1844970 99 + 9036312 ---------AUGUUCUGUCCUUGGUUGGACGAGCUCGAUCCUUGCGAAUUAAUGCCUAAUGCUUGUAAGUUAGGCAUGUCCAAGGGUCCCCUGUUCAUCCAGCCGCCA--------- ---------.............(((((((.((((..((((((((.((....(((((((((........))))))))).))))))))))....)))).)))))))....--------- ( -41.90, z-score = -5.82, R) >droWil1.scaffold_180701 2644230 99 - 3904529 ---------AUAUUUUGACCUUGAUUCGAUGAGCUCGAUCCUUGCGAAUUGAUGCCUAAUGCCUGCAAAUUGGGCAUAUCCAAAUGUCCCCUGUUCAUCCAACCGCCA--------- ---------..................(((((((..(((..(((.((....(((((((((........))))))))).)))))..)))....))))))).........--------- ( -23.50, z-score = -2.06, R) >droVir3.scaffold_12875 8749820 99 - 20611582 ---------AUGUUUUGCCCUUGAUUGGAAGAGCUGGAUCCUUGCGAAUUUAUGCCUAAUGCUUGUAGGUUAGGCAUGUCCAAAUGUCCCCUGUUCAUCCAACCACCA--------- ---------............((.(((((.((((.((((..(((.((...((((((((((........)))))))))))))))..))))...)))).))))).))...--------- ( -29.10, z-score = -2.52, R) >droMoj3.scaffold_6496 11860944 99 - 26866924 ---------AUGUUUUGCCCUUGAUUUGAAGAGCUGGAUCCUUGCGAAUUUAUGCCUAACGCUUGUAGGUUAGGCAUGUCCAAAUGUCCCCUGUUCAUCCAACCGCUA--------- ---------.......((..(((...((((.((..((((..(((.((...((((((((((........)))))))))))))))..)))).)).))))..)))..))..--------- ( -28.00, z-score = -2.84, R) >droGri2.scaffold_15245 11705200 99 + 18325388 ---------AUGUUUUGCCCUUGAUUUGAAGAGCUGGAUCCUUGAGAAUUUAUGCCUAACGCUUGUAGGUUAGGCAUGUCCAAAUGUCCCCUGUUCAUCCAACCGCCA--------- ---------.......((..(((...((((.((..((((..(((.((...((((((((((........)))))))))))))))..)))).)).))))..)))..))..--------- ( -27.60, z-score = -2.67, R) >anoGam1.chr2R 22228970 114 - 62725911 ACGCCCGCACCAUUACUCCCCGGACCGCCCGGUGCCGAUCCGCCGGCACCGCCGCUUCCACUCAGAUGGUUCCCCAUCCCUGCGUGGUGCCCGCCGUGCAGGCCCCCACCGACG--- ..(((.((((..........((((((....))).)))....((.((((((((.((.........(((((....)))))...)))))))))).)).)))).)))...........--- ( -42.80, z-score = -2.26, R) >apiMel3.GroupUn 25246352 105 + 399230636 ------------AUACCACCACCACCGCCGCCGCCAUUAUUCUGCCCGGUAAUGCCCAAUGCUUGAAGGUUAGGCAUAUCCAAAUUGCCUCGAUUCCCUGGAUUGUUCCACCCAUUU ------------...................................((((((.....((((((((...))))))))......)))))).........((((....))))....... ( -15.70, z-score = 1.62, R) >consensus _________AUGUUCUGGCCUUGAUUGGAGGAGCUCGAUCCCUGCGAAUUGAUGCCUAACGCCUGCAGGUUAGGCAUGUCCAAGUGGCCCCUGUUCAUCCAGCCGCUA_________ .....................((.((.((.((((.......(((.((....(((((((((........))))))))).))))).........)))).)).)).))............ (-12.86 = -12.63 + -0.23)

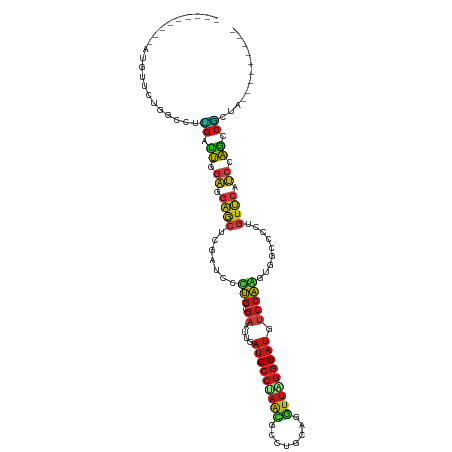
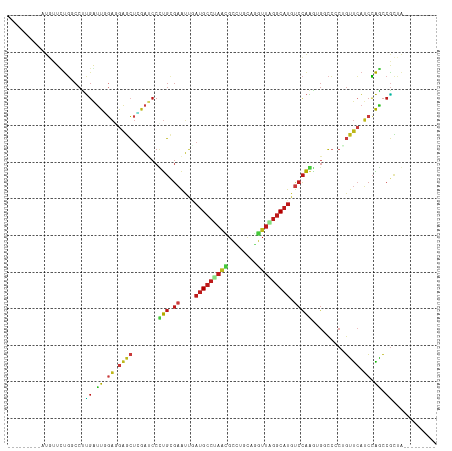
| Location | 19,747,561 – 19,747,662 |
|---|---|
| Length | 101 |
| Sequences | 14 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 71.34 |
| Shannon entropy | 0.62484 |
| G+C content | 0.57902 |
| Mean single sequence MFE | -35.63 |
| Consensus MFE | -18.53 |
| Energy contribution | -17.55 |
| Covariance contribution | -0.98 |
| Combinations/Pair | 1.82 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.942909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19747561 101 - 21146708 ------GAACCAGCC---GCAUAACGGCAACGUGGGCGGCAGCAACGGCUGGAUGAACCGUGGCCACCUGGACAUGCCCAACCUGCAGGCCCUGGGCAUCAACUCGCAGG ------......(((---((...(((....)))..)))))......((((((.....))..)))).((((((.(((((((.((....))...)))))))....)).)))) ( -40.30, z-score = -0.62, R) >droSim1.chr2R 18311047 101 - 19596830 ------GAACCAGCC---GCAUAACGGCAACGUGGGCGGCAGCAACGGCUGGAUGAACCGUGGCCACCUGGACAUGCCCAACCUGCAGGCGCUGGGCAUCAACUCGCAGG ------......(((---((...(((....)))..)))))......((((((.....))..)))).((((((.(((((((.((....))...)))))))....)).)))) ( -40.30, z-score = -0.60, R) >droSec1.super_9 3039057 101 - 3197100 ------GAACCAGCC---GCAUAACGGCAACGUGGGCGGCAGCAACGGCUGGAUGAACCGUGGCCACCUGGACAUGCCCAACCUGCAGGCGCUGGGCAUCAACUCGCAGG ------......(((---((...(((....)))..)))))......((((((.....))..)))).((((((.(((((((.((....))...)))))))....)).)))) ( -40.30, z-score = -0.60, R) >droYak2.chr2R 19730145 101 - 21139217 ------GAACCAGCC---CCACAACGGCAACGUGGGCGGCAGCAACGGCUGGAUGAACCGCGGCCACCUGGACAUGCCCAACCUGCAGGCGCUGGGCAUCAAUUCGCAGG ------......(((---((...(((....))).)).)))......((((((......).))))).((((((.(((((((.((....))...)))))))....)).)))) ( -38.40, z-score = -0.17, R) >droEre2.scaffold_4845 21125189 101 - 22589142 ------GAACCAGCC---UCACAACGGGAACGUGGGCGGCAGCAACGGCUGGAUGAACCGUGGCCCCCUGGACAUGCCUAACCUGCAGGCGCUGGGCAUCAAUUCGCAGG ------...(((((.---((.(((((....)))(((.(((....((((.........)))).))))))))))...((((.......)))))))))((........))... ( -33.20, z-score = 1.26, R) >droAna3.scaffold_13266 426443 101 - 19884421 ------GAACCAGCC---GCACAACGGCAACGUGGGCGGCAGUAACGGCUGGAUGAACAGGGGUCACUUGGAUAUGCCGAACUUGCAGGCGCUAGGCAUCAAUUCGCAGG ------((.((.(((---((...(((....)))..)))))........(((......))))).)).((((((.(((((......((....))..)))))....)).)))) ( -33.50, z-score = -0.77, R) >dp4.Unknown_group_152 9413 98 - 20572 ------GAACCAGCC---CCACAAUGGAAACGU---AGGCAGUGGCGGCUGGAUGAACAGGGGACCCUUGGACAUGCCUAACUUACAAGCAUUAGGCAUUAAUUCGCAAG ------(((((((((---((((.(((....)))---.....)))).))))))(((..(((((...)))))..)))((((((..........)))))).....)))..... ( -34.10, z-score = -2.77, R) >droPer1.super_2 1845001 98 - 9036312 ------GAACCAGCC---CCACAAUGGAAACGU---AGGCAGUGGCGGCUGGAUGAACAGGGGACCCUUGGACAUGCCUAACUUACAAGCAUUAGGCAUUAAUUCGCAAG ------(((((((((---((((.(((....)))---.....)))).))))))(((..(((((...)))))..)))((((((..........)))))).....)))..... ( -34.10, z-score = -2.77, R) >droWil1.scaffold_180701 2644261 104 + 3904529 ------GAAUCAACCGCAGCACAACGGCAACGUUGGCGGCAGUGGCGGUUGGAUGAACAGGGGACAUUUGGAUAUGCCCAAUUUGCAGGCAUUAGGCAUCAAUUCGCAAG ------(((((((((((.((.(((((....)))))...))....)))))))((((....(....)........(((((.........)))))....)))).))))..... ( -34.70, z-score = -2.24, R) >droVir3.scaffold_12875 8749851 98 + 20611582 ------GAACCAGCC---ACACAAUGGCAACGU---AGGCAAUGGUGGUUGGAUGAACAGGGGACAUUUGGACAUGCCUAACCUACAAGCAUUAGGCAUAAAUUCGCAAG ------...((((((---((.((.((.(.....---.).)).))))))))))..(((..(....)........((((((((..........))))))))...)))..... ( -28.60, z-score = -2.15, R) >droMoj3.scaffold_6496 11860975 98 + 26866924 ------GAACCAGCC---ACACAAUGGCAACGU---AGGCAAUAGCGGUUGGAUGAACAGGGGACAUUUGGACAUGCCUAACCUACAAGCGUUAGGCAUAAAUUCGCAAG ------...((((((---...(.(((....)))---.)((....))))))))..(((..(....)........((((((((((.....).)))))))))...)))..... ( -29.50, z-score = -2.74, R) >droGri2.scaffold_15245 11705231 98 - 18325388 ------GAAUCAGCC---ACACAAUGGCAACGU---AGGCAAUGGCGGUUGGAUGAACAGGGGACAUUUGGACAUGCCUAACCUACAAGCGUUAGGCAUAAAUUCUCAAG ------...((((((---...(.(((....)))---.)((....))))))))........((((.((((....((((((((((.....).)))))))))))))))))... ( -26.80, z-score = -1.75, R) >anoGam1.chr2R 22229010 110 + 62725911 CAACUCGAGCGGCCCAAACGGGCAGCGCGGGGGAGGCGUCGGUGGGGGCCUGCACGGCGGGCACCACGCAGGGAUGGGGAACCAUCUGAGUGGAAGCGGCGGUGCCGGCG ...((((.((.((((....)))).)).))))(.((((.((....)).)))).)...((.((((((.(((..((((((....))))))..)))(......))))))).)). ( -56.20, z-score = -2.36, R) >apiMel3.GroupUn 25246380 104 - 399230636 ------GGGCCAUUCAGGUAAUAAUAUGGGUCCAAAUGGGUGGAACAAUCCAGGGAAUCGAGGCAAUUUGGAUAUGCCUAACCUUCAAGCAUUGGGCAUUACCGGGCAGA ------..(((.....((((((.....((.(((...(((((......))))).)))....(((((.........)))))..)).....((.....)))))))).)))... ( -28.80, z-score = -0.02, R) >consensus ______GAACCAGCC___GCACAACGGCAACGUGGGCGGCAGUAACGGCUGGAUGAACAGGGGCCACUUGGACAUGCCUAACCUGCAGGCGUUAGGCAUCAAUUCGCAGG .........((((((........(((....))).............))))))..............((((((.(((((((............)))))))....)).)))) (-18.53 = -17.55 + -0.98)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:46:44 2011