| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,689,482 – 19,689,598 |
| Length | 116 |
| Max. P | 0.960870 |

| Location | 19,689,482 – 19,689,598 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 69.57 |
| Shannon entropy | 0.57756 |
| G+C content | 0.48919 |
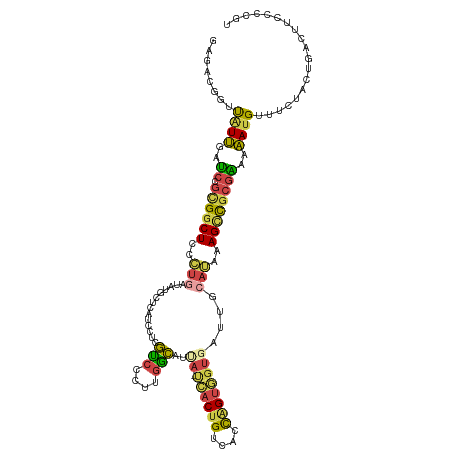
| Mean single sequence MFE | -34.63 |
| Consensus MFE | -14.62 |
| Energy contribution | -13.97 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.67 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.960870 |
| Prediction | RNA |
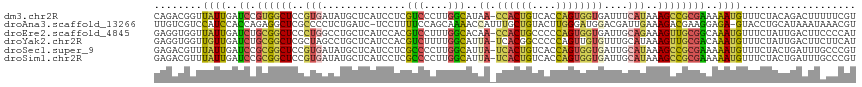
Download alignment: ClustalW | MAF
>dm3.chr2R 19689482 116 - 21146708 CAGACGGUUAUUGAUCCGUGGCUCCGUGAUAUGCUCAUCCUCGUCCCUUGGCAUAA-CCACUGUCACCAGUGGUGAUUUCAUAAAGCCGCGAAAAAUGUUUCUACAGACUUUUUCGU ...((((........))))((((..(((((((((.((...........)))))))(-((((((....)))))))....))))..))))((((((((.(((......))))))))))) ( -36.40, z-score = -3.43, R) >droAna3.scaffold_13266 366989 115 - 19884421 UUGUCGUCCAUCCACCAGAGGCUCGCCCCUCUGAUC-UCCUUUUCCAGCAAAACCAUUUGCUGUACUUGGGAUGGACGAUUGAAAGACGAAGGAGA-GUACCUGCAUAAAUAAACGU ..(((((((((((..((((((......))))))...-........(((((((....)))))))......)))))))))))........(.(((...-...))).)............ ( -37.70, z-score = -3.40, R) >droEre2.scaffold_4845 21049720 116 - 22589142 GAGGUGGUUAUUGAUCUGCGGCUCCCUGGCCUGCUCAUCCACGUCCUUUGGCACAA-CCACUGCCCCCAGUGGUGAUUGCAGAAAGUUGCGGCAAAUGUUUCUAUUGACUUCCCCAU ((.((((....(((...((((((....)))).))))).)))).))((((.(((..(-((((((....)))))))...)))..))))....((..((.(((......)))))..)).. ( -32.40, z-score = 0.21, R) >droYak2.chr2R 19671555 116 - 21139217 GAGGUGGUUGUUGAUCUGCGGCUCGCUAGCCUGCUCAUCCACGUCUUUUGGCAUUA-UCACGGCCCCCAGUUGUGUUUGCAUAAAGUUGCGACAAAUGUUUCUAUUGACUUCUUCAU ((((.((((((.(((..((((((....)))).))..))).)).......((((((.-.((((((.....)))))).(((((......)))))..))))))......)))).)))).. ( -29.10, z-score = -0.13, R) >droSec1.super_9 2979521 116 - 3197100 GAGACGUUUAUUGAUCCGCGGCUCCGUGAUAUGCUCAUCCUCGCCCCUUGGCAUUA-UCACUGUCACCAGUGGUGAUUGCAUAAAGCCGCGAAAAAUGUUUCUACUGAUUUGCCCGU (((((((((.......(((((((......(((((........(((....)))((((-((((((....)))))))))).))))).)))))))..)))))))))............... ( -36.10, z-score = -2.96, R) >droSim1.chr2R 18254948 116 - 19596830 GAGACGUUUAUUGAUCCGCGGCUCCGUGAUAUGCUCAUCCUCGCCCCUUGGCAUUA-UCACUGUCACCAGUGGUGAUUGCAUAAAGCCGCGAAAAAUGUUUCUACUGAUUUGCCCGU (((((((((.......(((((((......(((((........(((....)))((((-((((((....)))))))))).))))).)))))))..)))))))))............... ( -36.10, z-score = -2.96, R) >consensus GAGACGGUUAUUGAUCCGCGGCUCCCUGAUAUGCUCAUCCUCGUCCCUUGGCAUUA_UCACUGUCACCAGUGGUGAUUGCAUAAAGCCGCGAAAAAUGUUUCUACUGACUUCCCCGU ........((((..((.((((((..(((..............(((....))).....((((((....))))))......)))..))))))))..))))................... (-14.62 = -13.97 + -0.66)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:46:37 2011