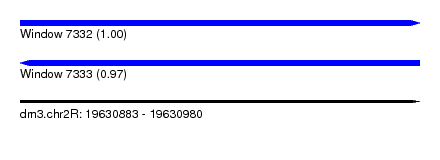
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,630,883 – 19,630,980 |
| Length | 97 |
| Max. P | 0.998222 |

| Location | 19,630,883 – 19,630,980 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 67.14 |
| Shannon entropy | 0.57906 |
| G+C content | 0.46904 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -13.17 |
| Energy contribution | -14.27 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
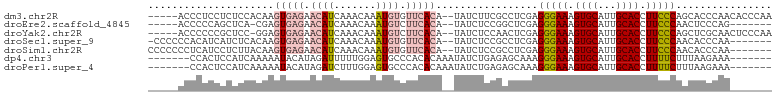
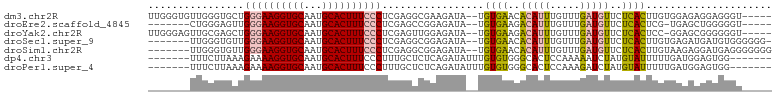
>dm3.chr2R 19630883 97 + 21146708 -----ACCCUCCUCUCCACAAGUGAGAACAUCAAACAAAUGUGUUCACA--UAUCUUCGCCUCGAGGGAAAGUGCAUUGCACCUUCCCAGCACCCAACACCCAA -----................(((((.((((.......)))).))))).--..............(((((.((((...)))).)))))................ ( -19.90, z-score = -2.09, R) >droEre2.scaffold_4845 20994029 89 + 22589142 -----ACCCCCAGCUCA-CGAGUGAGAACAUCAAACAAAUGUCUUCACA--UAUCUCCGGCUCGAGGGAAAGUGCAUUGCACCUUCCCAACUCCCAG------- -----....(.((((..-.((((((..((((.......))))..)))).--..))...)))).).(((((.((((...)))).))))).........------- ( -22.10, z-score = -2.38, R) >droYak2.chr2R 19609028 96 + 21139217 -----ACCCCCCGCUCC-GGAGUGAGAACAUCAAACAAAUGUCUUCACA--UAUCUCCAACUCGAGGGAAAGUGCAUUGCACCUUCCCAGCUCGCAACUCCCAA -----.......(((..-(((((((..((((.......))))..)))).--....))).......(((((.((((...)))).))))))))............. ( -23.70, z-score = -2.51, R) >droSec1.super_9 2911094 94 + 3197100 -CCCCCCACAUCAUCUCACAAGUGAGAACAUCAAACAAAUGUGUUCACA--UAUCUCCGCCUCGAGGGAAAGUGCAUUGCACCUUCCCAACACCCAA------- -............(((((....))))).............(((((....--..((........))(((((.((((...)))).))))))))))....------- ( -20.50, z-score = -2.82, R) >droSim1.chr2R 18197137 95 + 19596830 CCCCCCCUCAUCCUCUUACAAGUGAGAACAUCAAACAAAUGUGUUCACA--UAUCUCCGCCUCGAGGGAAAGUGCAUUGCACCUUCCCAACACCCAA------- .....................(((((.((((.......)))).))))).--..............(((((.((((...)))).))))).........------- ( -19.90, z-score = -2.82, R) >dp4.chr3 10413701 90 - 19779522 -------CCACUCCAUCAAAAAUACAUAGAUUUUUGGAGUGCCCACACAAAUAUCUGAGAGCAAAGGGAAAGUGCAUUGCACCUUUUCUUUAAGAAA------- -------.(((((((...(((((......)))))))))))).....................((((..((.((((...)))).))..))))......------- ( -19.40, z-score = -1.62, R) >droPer1.super_4 2473676 90 - 7162766 -------CCACUCCAUCAAAAAUACAUAGAUCUUUGGAGUGCCCACACAAAUAUCUGAGAGCAAAGGGAAAGUGCAUUGCACCUUUUCUUUAAGAAA------- -------.(((((((...................))))))).....................((((..((.((((...)))).))..))))......------- ( -18.51, z-score = -1.17, R) >consensus _____ACCCACCCCUUCACAAGUGAGAACAUCAAACAAAUGUCUUCACA__UAUCUCCGACUCGAGGGAAAGUGCAUUGCACCUUCCCAACACCCAA_______ .....................(((((.((((.......)))).))))).................(((((.((((...)))).)))))................ (-13.17 = -14.27 + 1.11)
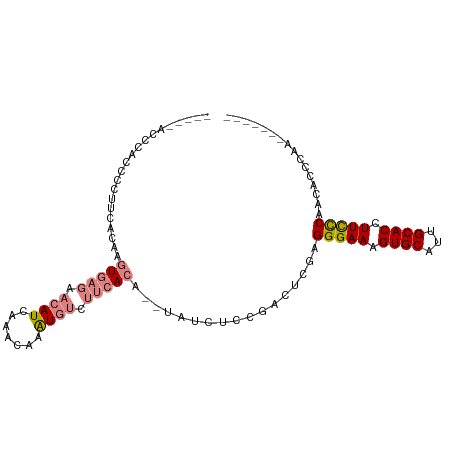
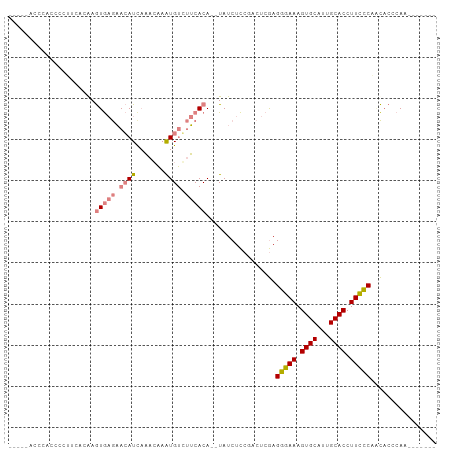
| Location | 19,630,883 – 19,630,980 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 67.14 |
| Shannon entropy | 0.57906 |
| G+C content | 0.46904 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -17.37 |
| Energy contribution | -17.37 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
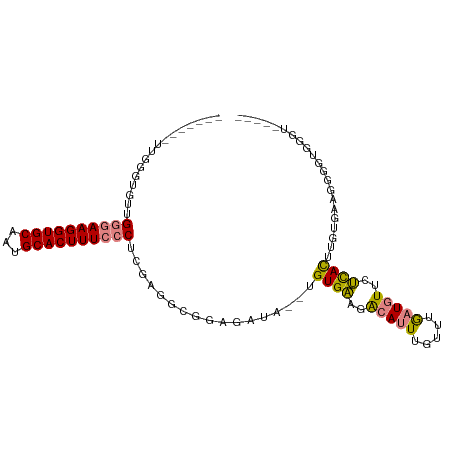
>dm3.chr2R 19630883 97 - 21146708 UUGGGUGUUGGGUGCUGGGAAGGUGCAAUGCACUUUCCCUCGAGGCGAAGAUA--UGUGAACACAUUUGUUUGAUGUUCUCACUUGUGGAGAGGAGGGU----- (((.(((((...((((((((((((((...))))))))))....))))..))))--).)))..(((((.....)))))((((.(((.....)))))))..----- ( -29.10, z-score = -0.91, R) >droEre2.scaffold_4845 20994029 89 - 22589142 -------CUGGGAGUUGGGAAGGUGCAAUGCACUUUCCCUCGAGCCGGAGAUA--UGUGAAGACAUUUGUUUGAUGUUCUCACUCG-UGAGCUGGGGGU----- -------.........((((((((((...)))))))))).....((((.....--.((((..(((((.....)))))..))))...-....))))....----- ( -31.42, z-score = -1.81, R) >droYak2.chr2R 19609028 96 - 21139217 UUGGGAGUUGCGAGCUGGGAAGGUGCAAUGCACUUUCCCUCGAGUUGGAGAUA--UGUGAAGACAUUUGUUUGAUGUUCUCACUCC-GGAGCGGGGGGU----- ((.(((((.(.((((.((((((((((...))))))))))....((..((.(.(--(((....)))).).))..)))))).))))))-.)).........----- ( -34.30, z-score = -1.92, R) >droSec1.super_9 2911094 94 - 3197100 -------UUGGGUGUUGGGAAGGUGCAAUGCACUUUCCCUCGAGGCGGAGAUA--UGUGAACACAUUUGUUUGAUGUUCUCACUUGUGAGAUGAUGUGGGGGG- -------(((.(((((((((((((((...))))))))))((......))))))--).))).((((((..........(((((....))))).)))))).....- ( -27.40, z-score = -0.92, R) >droSim1.chr2R 18197137 95 - 19596830 -------UUGGGUGUUGGGAAGGUGCAAUGCACUUUCCCUCGAGGCGGAGAUA--UGUGAACACAUUUGUUUGAUGUUCUCACUUGUAAGAGGAUGAGGGGGGG -------....((((((........)))))).((..(((((....((((.(.(--(((....)))).).))))..((((((........)))))))))))..)) ( -27.80, z-score = -1.17, R) >dp4.chr3 10413701 90 + 19779522 -------UUUCUUAAAGAAAAGGUGCAAUGCACUUUCCCUUUGCUCUCAGAUAUUUGUGUGGGCACUCCAAAAAUCUAUGUAUUUUUGAUGGAGUGG------- -------......((((..(((((((...)))))))..))))((((.((.(....).)).))))(((((((((((......)))))...))))))..------- ( -22.30, z-score = -1.53, R) >droPer1.super_4 2473676 90 + 7162766 -------UUUCUUAAAGAAAAGGUGCAAUGCACUUUCCCUUUGCUCUCAGAUAUUUGUGUGGGCACUCCAAAGAUCUAUGUAUUUUUGAUGGAGUGG------- -------......((((..(((((((...)))))))..))))((((.((.(....).)).))))(((((((((((......)))))...))))))..------- ( -22.40, z-score = -1.15, R) >consensus _______UUGGGUGUUGGGAAGGUGCAAUGCACUUUCCCUCGAGGCGGAGAUA__UGUGAAGACAUUUGUUUGAUGUUCUCACUUGUGAAGGGGUGGGU_____ ................((((((((((...)))))))))).................((((..(((((.....)))))..))))..................... (-17.37 = -17.37 + 0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:46:30 2011