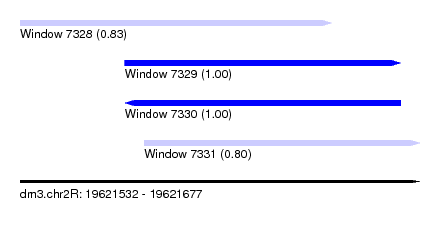
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,621,532 – 19,621,677 |
| Length | 145 |
| Max. P | 0.999449 |

| Location | 19,621,532 – 19,621,645 |
|---|---|
| Length | 113 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.70 |
| Shannon entropy | 0.45775 |
| G+C content | 0.29531 |
| Mean single sequence MFE | -21.56 |
| Consensus MFE | -11.70 |
| Energy contribution | -11.55 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.831760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
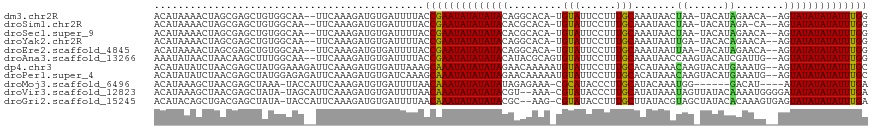
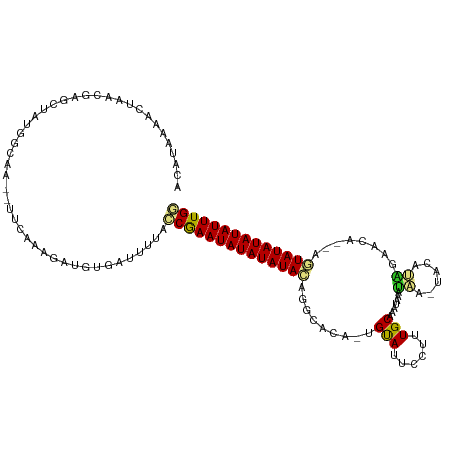
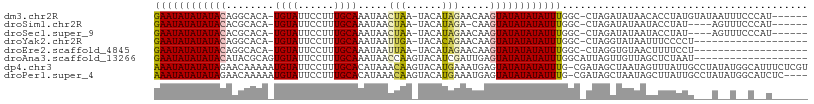
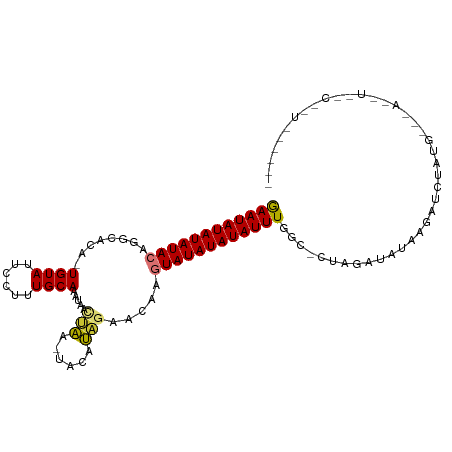
>dm3.chr2R 19621532 113 + 21146708 ACAUAAAACUAGCGAGCUGUGGCAA--UUCAAAGAUGUGAUUUUACCGAAUAUAUAUACAGGCACA-UGUAUUCCUUUGCAAAUAACUAA-UACAUAGAACA--AGUAUAUAUAUUUGG ...(((((...(((..((.(((...--.))).)).)))..)))))((((((((((((((..(..((-((((((..((.......))..))-))))).)..).--.)))))))))))))) ( -20.10, z-score = -0.85, R) >droSim1.chr2R 18187636 112 + 19596830 ACAUAAAACUAGCGAGCUGUGGCAA--UUCAAAGAUGUGAUUUUACCGAAUAUAUAUACACGCACA-UGUAUUCCUUUGCAAAUAACUAA-UACAUAGA-CA--AGUAUAUAUAUUUGG ...(((((...(((..((.(((...--.))).)).)))..)))))((((((((((((((......(-((((((..((.......))..))-)))))...-..--.)))))))))))))) ( -19.90, z-score = -1.06, R) >droSec1.super_9 2901898 113 + 3197100 ACAUAAAACUAGCGAGCUGUGGCAA--UUCAAAGAUGUGAUUUUACCGAAUAUAUAUACACGCACA-UGUAUUCCUUUGCAAAUAACUAA-UACAUAGAACA--AGUAUAUAUAUUUGG ...(((((...(((..((.(((...--.))).)).)))..)))))((((((((((((((.......-((((......)))).....(((.-....)))....--.)))))))))))))) ( -19.90, z-score = -1.06, R) >droYak2.chr2R 19599584 113 + 21139217 ACAUAAAACUAGCGAGCUGUGGCAA--UUCAAAGAUGUGAUUUUACCGAAUAUAUAUACAGGCACA-UGUAUUCCUUUGCAAAUAAUUGA-UACACAGAACA--AGUAUAUAUAUUUGG ...(((((...(((..((.(((...--.))).)).)))..)))))((((((((((((((.......-((((......)))).....(((.-....)))....--.)))))))))))))) ( -19.00, z-score = -0.25, R) >droEre2.scaffold_4845 20984371 113 + 22589142 ACAUAAAACUAGCGAGCUGUGGCAA--UUCAAAGAUGUGAUUUUACCGAAUAUAUAUACAGGCACA-UGUAUUCCUUUGCAAAUAAUUAA-UACAUAGAACA--AGUAUAUAUAUUUGG ...(((((...(((..((.(((...--.))).)).)))..)))))((((((((((((((..(..((-((((((..((.......))..))-))))).)..).--.)))))))))))))) ( -20.10, z-score = -0.85, R) >droAna3.scaffold_13266 8260376 115 + 19884421 AAAUAUAACUAACAAGCUUUGGCAA--UUCAAAGAUGUGAUUUUACCGAAUAUAUAUACAUACGCAGUGUAUUCCUUUGCAAAUAACCAAGUACAUCGAUUG--AGUAUAUAUAUUUGG ............((..((((((...--.))))))...))......((((((((((((((.....(.(((((((................))))))).)....--.)))))))))))))) ( -23.89, z-score = -2.36, R) >dp4.chr3 10402847 117 - 19779522 ACAUAUAUCUAACGAGCUAUGGAAAGAUUCAAAGAUGUGAUUAAAGCAAAUAUAUAUAGAACAAAAAUGUAUUCCUUUGCACAUAAACAAGUACAUGAAAUG--AGUAUAUAUAUUUGC ...(((((((...(((((......)).)))..)))))))......(((((((((((((........(((((((..((((....))))..)))))))......--..))))))))))))) ( -23.29, z-score = -2.71, R) >droPer1.super_4 2462874 117 - 7162766 ACAUAUAUCUAACGAGCUAUGGAGAGAUUCAAAGAUGUGAUCAAAGCAAAUAUAUAUAGAACAAAAAUGUAUUCCUUUGCACAUAAACAAGUACAUGAAAUG--AGUAUAUAUAUUUGC .(((((.((....)).((.((((....)))).)))))))......(((((((((((((........(((((((..((((....))))..)))))))......--..))))))))))))) ( -23.89, z-score = -2.55, R) >droMoj3.scaffold_6496 26548690 107 - 26866924 ACAUAAAGCUAACGAGCUAAA-UACCAUUCAAAGAUGUGAUUUUAACAAAUAUAUAUAUAGAGAAA-CGCAUACCCUUGCAUACAAAUGG------GACAU----AUAUAUAUAUUUGA ......((((....))))(((-(..((((....))))..))))...((((((((((((((......-.(((......)))...(....).------....)----))))))))))))). ( -18.70, z-score = -2.67, R) >droVir3.scaffold_12823 1955316 115 - 2474545 ACAUAAAGCUAACGAGCUAUA-UAGCAUUCAAAGAUGUGAUUUUAACAAAUAUAUAUACGU--AAA-CGUAUACCCUUGCAUAUAAAUAGUUAUACAAAAUGGGGAUAUAUAUAUUUGA ..(((.((((....)))).))-).(((((....)))))........((((((((((((((.--...-))))..((((....(((((....)))))......))))...)))))))))). ( -22.40, z-score = -2.09, R) >droGri2.scaffold_15245 6157999 115 + 18325388 ACAUACAGCUGACGAGCUAUA-UACCAUUCAAAGAUGUGAUUUUAACAAAUAUAUAUACGC--AAG-CGUAUACCUUUGCUUAUACGUAGCUAUACACAAAGUGAGUAUAUAUAUUUGA ..(((.((((....)))).))-).(((((....)))).).......(((((((((((((((--..(-((((((........))))))).))....(((...))).))))))))))))). ( -26.00, z-score = -2.52, R) >consensus ACAUAAAACUAACGAGCUAUGGCAA__UUCAAAGAUGUGAUUUUACCGAAUAUAUAUACAGGCACA_UGUAUUCCUUUGCAAAUAAAUAA_UACAUAGAACA__AGUAUAUAUAUUUGG .............................................((((((((((((((.........(((......))).......((......))........)))))))))))))) (-11.70 = -11.55 + -0.15)
| Location | 19,621,570 – 19,621,670 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 68.57 |
| Shannon entropy | 0.60538 |
| G+C content | 0.27981 |
| Mean single sequence MFE | -21.01 |
| Consensus MFE | -14.81 |
| Energy contribution | -14.32 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999449 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 19621570 100 + 21146708 UUUUACCGAAUAUAUAUACAGGCACA-UGUAUUCCUUUGCAAAUAACUAA---UACAUAGAACAAGUAUAUAUAUUUGGCCU-AGAUAUAACACCUAUGUAUAAU------------ .....((((((((((((((..(..((-((((((..((.......))..))---))))).)..)..))))))))))))))...-..(((((.....))))).....------------ ( -19.00, z-score = -2.34, R) >droSim1.chr2R 18187674 95 + 19596830 UUUUACCGAAUAUAUAUACACGCACA-UGUAUUCCUUUGCAAAUAACUAA---UACAUAGA-CAAGUAUAUAUAUUUGGCCU-AGAUAUAAUACCUAUAGU---------------- .....((((((((((((((......(-((((((..((.......))..))---)))))...-...))))))))))))))...-..................---------------- ( -18.50, z-score = -2.89, R) >droSec1.super_9 2901936 96 + 3197100 UUUUACCGAAUAUAUAUACACGCACA-UGUAUUCCUUUGCAAAUAACUAA---UACAUAGAACAAGUAUAUAUAUUUGGCCU-AGAUAUAAUACCUAUAGU---------------- .....((((((((((((((.......-((((......)))).....(((.---....))).....))))))))))))))...-..................---------------- ( -18.50, z-score = -2.86, R) >droYak2.chr2R 19599622 87 + 21139217 UUUUACCGAAUAUAUAUACAGGCACA-UGUAUUCCUUUGCAAAUAAUUGA---UACACAGAACAAGUAUAUAUAUUUGGCCU-AGGUAUAAU------------------------- .....((((((((((((((.......-((((......)))).....(((.---....))).....))))))))))))))...-.........------------------------- ( -17.60, z-score = -2.12, R) >droEre2.scaffold_4845 20984409 87 + 22589142 UUUUACCGAAUAUAUAUACAGGCACA-UGUAUUCCUUUGCAAAUAAUUAA---UACAUAGAACAAGUAUAUAUAUUUGGCCU-AGGUGUAAC------------------------- .....((((((((((((((..(..((-((((((..((.......))..))---))))).)..)..))))))))))))))...-.........------------------------- ( -18.70, z-score = -2.18, R) >droAna3.scaffold_13266 8260414 90 + 19884421 UUUUACCGAAUAUAUAUACAUACGCAGUGUAUUCCUUUGCAAAUAACCAA--GUACAUCGAUUGAGUAUAUAUAUUUGGCAUUAGUUGUUAG------------------------- .....((((((((((((((.....(.(((((((................)--)))))).).....)))))))))))))).............------------------------- ( -19.29, z-score = -2.43, R) >dp4.chr3 10402887 108 - 19779522 UUAAAGCAAAUAUAUAUAGAACAAAAAUGUAUUCCUUUGCACAUAAACAA--GUACAUGAAAUGAGUAUAUAUAUUUGCGAUAGCUAAUAGUUUAUUGCCUAUAUGGCAU------- .....(((((((((((((........(((((((..((((....))))..)--))))))........))))))))))))).................((((.....)))).------- ( -24.99, z-score = -2.84, R) >droPer1.super_4 2462914 108 - 7162766 UCAAAGCAAAUAUAUAUAGAACAAAAAUGUAUUCCUUUGCACAUAAACAA--GUACAUGAAAUGAGUAUAUAUAUUUGCGAUAGCUAAUAGCUUAUUGCCUAUAUGGCAU------- .....(((((((((((((........(((((((..((((....))))..)--))))))........)))))))))))))...(((.....)))...((((.....)))).------- ( -26.39, z-score = -3.01, R) >droGri2.scaffold_15245 6158038 113 + 18325388 UUUUAACAAAUAUAUAUACGCA---AGCGUAUACCUUUGCUUAUACGUAGCUAUACACAAAGUGAGUAUAUAUAUUUGAUAU-AGUUAUAAGUAGUUAGUUGUAAGACUCCUGGCAA ......(((((((((((((((.---.(((((((........))))))).))....(((...))).)))))))))))))....-...........(((((..(.....)..))))).. ( -26.10, z-score = -1.58, R) >consensus UUUUACCGAAUAUAUAUACACGCACA_UGUAUUCCUUUGCAAAUAACUAA___UACAUAGAACAAGUAUAUAUAUUUGGCCU_AGUUAUAAU___U____U________________ .....((((((((((((((........((((.....(((........)))...))))........))))))))))))))...................................... (-14.81 = -14.32 + -0.49)

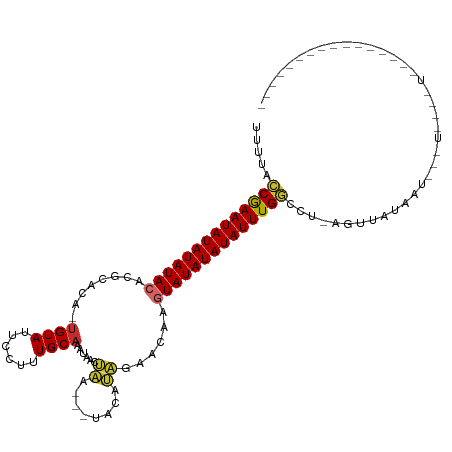
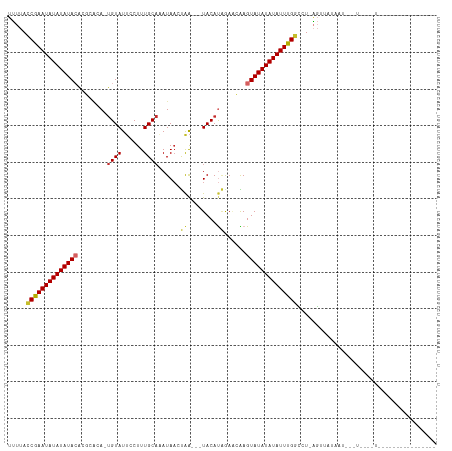
| Location | 19,621,570 – 19,621,670 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 68.57 |
| Shannon entropy | 0.60538 |
| G+C content | 0.27981 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -11.30 |
| Energy contribution | -11.38 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.995291 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 19621570 100 - 21146708 ------------AUUAUACAUAGGUGUUAUAUCU-AGGCCAAAUAUAUAUACUUGUUCUAUGUA---UUAGUUAUUUGCAAAGGAAUACA-UGUGCCUGUAUAUAUAUUCGGUAAAA ------------........((((((...)))))-).(((.(((((((((((..((..((((((---((..((.......))..))))))-)).))..))))))))))).))).... ( -22.70, z-score = -1.40, R) >droSim1.chr2R 18187674 95 - 19596830 ----------------ACUAUAGGUAUUAUAUCU-AGGCCAAAUAUAUAUACUUG-UCUAUGUA---UUAGUUAUUUGCAAAGGAAUACA-UGUGCGUGUAUAUAUAUUCGGUAAAA ----------------....((((((...)))))-).(((.(((((((((((.((-(.((((((---((..((.......))..))))))-)).))).))))))))))).))).... ( -25.30, z-score = -2.77, R) >droSec1.super_9 2901936 96 - 3197100 ----------------ACUAUAGGUAUUAUAUCU-AGGCCAAAUAUAUAUACUUGUUCUAUGUA---UUAGUUAUUUGCAAAGGAAUACA-UGUGCGUGUAUAUAUAUUCGGUAAAA ----------------....((((((...)))))-).(((.(((((((((((.(((..((((((---((..((.......))..))))))-)).))).))))))))))).))).... ( -23.80, z-score = -2.24, R) >droYak2.chr2R 19599622 87 - 21139217 -------------------------AUUAUACCU-AGGCCAAAUAUAUAUACUUGUUCUGUGUA---UCAAUUAUUUGCAAAGGAAUACA-UGUGCCUGUAUAUAUAUUCGGUAAAA -------------------------.........-..(((.(((((((((((..((...(((((---((((....))).......)))))-)..))..))))))))))).))).... ( -18.51, z-score = -1.45, R) >droEre2.scaffold_4845 20984409 87 - 22589142 -------------------------GUUACACCU-AGGCCAAAUAUAUAUACUUGUUCUAUGUA---UUAAUUAUUUGCAAAGGAAUACA-UGUGCCUGUAUAUAUAUUCGGUAAAA -------------------------.........-..(((.(((((((((((..((..((((((---((..((.......))..))))))-)).))..))))))))))).))).... ( -21.50, z-score = -2.57, R) >droAna3.scaffold_13266 8260414 90 - 19884421 -------------------------CUAACAACUAAUGCCAAAUAUAUAUACUCAAUCGAUGUAC--UUGGUUAUUUGCAAAGGAAUACACUGCGUAUGUAUAUAUAUUCGGUAAAA -------------------------...........((((.(((((((((((..((((((.....--))))))....(((..(.....)..)))....))))))))))).))))... ( -19.40, z-score = -2.40, R) >dp4.chr3 10402887 108 + 19779522 -------AUGCCAUAUAGGCAAUAAACUAUUAGCUAUCGCAAAUAUAUAUACUCAUUUCAUGUAC--UUGUUUAUGUGCAAAGGAAUACAUUUUUGUUCUAUAUAUAUUUGCUUUAA -------.((((.....)))).................((((((((((((..........(((((--........)))))..((((((......))))))))))))))))))..... ( -26.10, z-score = -3.17, R) >droPer1.super_4 2462914 108 + 7162766 -------AUGCCAUAUAGGCAAUAAGCUAUUAGCUAUCGCAAAUAUAUAUACUCAUUUCAUGUAC--UUGUUUAUGUGCAAAGGAAUACAUUUUUGUUCUAUAUAUAUUUGCUUUGA -------.((((.....))))...(((.....)))...((((((((((((..........(((((--........)))))..((((((......))))))))))))))))))..... ( -28.20, z-score = -2.90, R) >droGri2.scaffold_15245 6158038 113 - 18325388 UUGCCAGGAGUCUUACAACUAACUACUUAUAACU-AUAUCAAAUAUAUAUACUCACUUUGUGUAUAGCUACGUAUAAGCAAAGGUAUACGCU---UGCGUAUAUAUAUUUGUUAAAA ..................................-.((.(((((((((((((.((....(((((((.((..((....))..)).))))))).---)).))))))))))))).))... ( -23.30, z-score = -1.35, R) >consensus ________________A____A___AUUAUAACU_AGGCCAAAUAUAUAUACUUGUUCUAUGUA___UUAGUUAUUUGCAAAGGAAUACA_UGUGCCUGUAUAUAUAUUCGGUAAAA .....................................(((.(((((((((((........((((......................))))........))))))))))).))).... (-11.30 = -11.38 + 0.09)

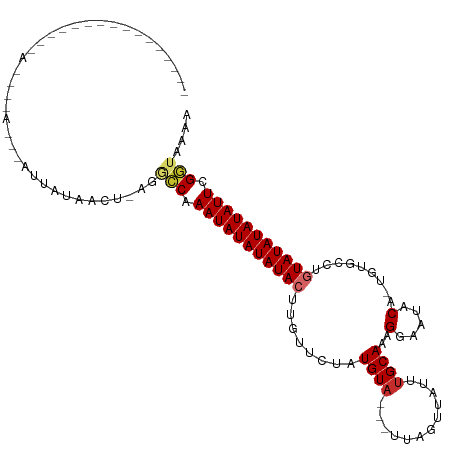
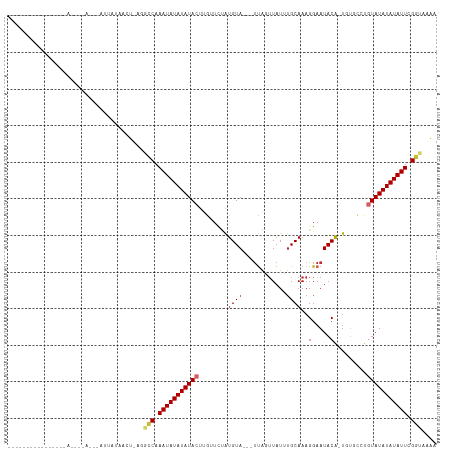
| Location | 19,621,577 – 19,621,677 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 71.61 |
| Shannon entropy | 0.53753 |
| G+C content | 0.28632 |
| Mean single sequence MFE | -16.13 |
| Consensus MFE | -10.29 |
| Energy contribution | -10.07 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.21 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.804134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19621577 100 + 21146708 GAAUAUAUAUACAGGCACA-UGUAUUCCUUUGCAAAUAACUAA-UACAUAGAACAAGUAUAUAUAUUUGGC-CUAGAUAUAACACCUAUGUAUAAUUUCCCAU------ (((..((((((.(((....-((((......))))........(-((..(((..((((((....))))))..-)))..)))....))).))))))..)))....------ ( -16.20, z-score = -1.15, R) >droSim1.chr2R 18187681 95 + 19596830 GAAUAUAUAUACACGCACA-UGUAUUCCUUUGCAAAUAACUAA-UACAUAGA-CAAGUAUAUAUAUUUGGC-CUAGAUAUAAUACCUAU----AGUUUCCCAU------ ((((((((((((......(-((((((..((.......))..))-)))))...-...))))))))))))((.-.....((((.....)))----).....))..------ ( -14.00, z-score = -1.08, R) >droSec1.super_9 2901943 96 + 3197100 GAAUAUAUAUACACGCACA-UGUAUUCCUUUGCAAAUAACUAA-UACAUAGAACAAGUAUAUAUAUUUGGC-CUAGAUAUAAUACCUAU----AGUUUCCCAU------ ((((((((((((.......-((((......)))).....(((.-....))).....))))))))))))((.-.....((((.....)))----).....))..------ ( -14.00, z-score = -1.14, R) >droYak2.chr2R 19599629 87 + 21139217 GAAUAUAUAUACAGGCACA-UGUAUUCCUUUGCAAAUAAUUGA-UACACAGAACAAGUAUAUAUAUUUGGC-CUAGGUAUAAUUUCCCCU------------------- (((..(((((..((((...-((((......)))).........-.........((((((....))))))))-))..)))))..)))....------------------- ( -15.40, z-score = -0.89, R) >droEre2.scaffold_4845 20984416 87 + 22589142 GAAUAUAUAUACAGGCACA-UGUAUUCCUUUGCAAAUAAUUAA-UACAUAGAACAAGUAUAUAUAUUUGGC-CUAGGUGUAACUUUUCCU------------------- .....(((((..((((..(-((((((..((.......))..))-)))))....((((((....))))))))-))..))))).........------------------- ( -15.50, z-score = -0.89, R) >droAna3.scaffold_13266 8260421 90 + 19884421 GAAUAUAUAUACAUACGCAGUGUAUUCCUUUGCAAAUAACCAAGUACAUCGAUUGAGUAUAUAUAUUUGGCAUUAGUUGUUAGCUCUAAU------------------- ((((((((((((.....(.(((((((................))))))).).....))))))))))))((((.....)))).........------------------- ( -14.89, z-score = -0.18, R) >dp4.chr3 10402894 108 - 19779522 AAAUAUAUAUAGAACAAAAAUGUAUUCCUUUGCACAUAAACAAGUACAUGAAAUGAGUAUAUAUAUUUG-CGAUAGCUAAUAGUUUAUUGCCUAUAUGGCAUUUCUCGU (((((((((((........(((((((..((((....))))..)))))))........)))))))))))(-(((.(((.....)))...((((.....))))....)))) ( -19.39, z-score = -0.88, R) >droPer1.super_4 2462921 104 - 7162766 AAAUAUAUAUAGAACAAAAAUGUAUUCCUUUGCACAUAAACAAGUACAUGAAAUGAGUAUAUAUAUUUG-CGAUAGCUAAUAGCUUAUUGCCUAUAUGGCAUCUC---- (((((((((((........(((((((..((((....))))..)))))))........))))))))))).-.(((.(((((((((.....).)))).)))))))..---- ( -19.69, z-score = -1.09, R) >consensus GAAUAUAUAUACAGGCACA_UGUAUUCCUUUGCAAAUAACUAA_UACAUAGAACAAGUAUAUAUAUUUGGC_CUAGAUAUAAGAUCUAUG___A__U__C__U______ ((((((((((((........((((......)))).....(((......))).....))))))))))))......................................... (-10.29 = -10.07 + -0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:46:29 2011