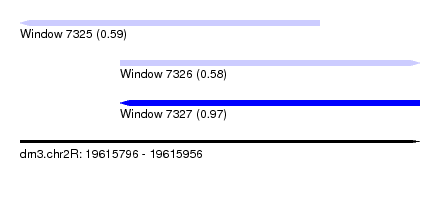
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,615,796 – 19,615,956 |
| Length | 160 |
| Max. P | 0.968775 |

| Location | 19,615,796 – 19,615,916 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.60 |
| Shannon entropy | 0.46422 |
| G+C content | 0.54043 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -22.21 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.594743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
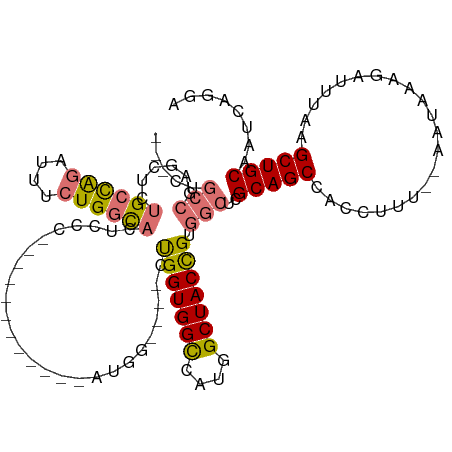
>dm3.chr2R 19615796 120 - 21146708 GUUCUGCCGAGCUCUGCCAGAUUUCUGGCACUCGCUUCCCGCCACCAAUGGCUGGCUGGUGGCCAUGGCUACCGUGGCUCGCAGCCCCCUUUUCAAUAAAGAUUUUAGCUGCAAUCAGGA .(((((..((((..((((((....))))))..(((..((.((((....)))).))..((((((....)))))))))))))(((((...((((.....))))......)))))...))))) ( -46.80, z-score = -2.30, R) >droSim1.chr2R 18181879 111 - 19596830 GCUCUGCCGAGCUCUGCCAGAUUUCUGGCACUCGGCUCCC-------AUGGCU--CUGGUGGCCAUGGCUACCGUGGCUCGCAGCCCCCCUUUCAAUAAAGAUUUGAGCUGCAAUCAGGA .....((((((...((((((....))))))))))))..((-------..((((--.(((((((....))))))).)))).(((((.......((((.......))))))))).....)). ( -47.51, z-score = -2.72, R) >droYak2.chr2R 19593728 95 - 21139217 ---GUUCCGAGUUUUGCUGGAUUUCUGGCACAUCA------------AUGG----CCGGUGGUCAUGGCUACUGUG----GCAGCCACCUGU--AAUAAGGAUUUAUGCUGCAAUCAAGA ---......(((...(((((((..(((((.((...------------.)))----))))..))).)))).)))(..----(((....(((..--....))).....)))..)........ ( -27.10, z-score = 0.12, R) >droEre2.scaffold_4845 20978629 99 - 22589142 ---GGGCGGCACUGUGCUGCAUUUCUGUUCCCACC------------AUCA----AUGGUGGCCAUGGCUACUGGGGCUCGCAGCCACCUUU--AGUAAAGAUUUAAGCUGCAAUCAGUA ---(((((((.....)))))..........(((((------------(...----.))))))))...((((..(((((.....))).))..)--)))..........((((....)))). ( -31.80, z-score = 0.02, R) >consensus ___CUGCCGAGCUCUGCCAGAUUUCUGGCACUCCC____________AUGG____CUGGUGGCCAUGGCUACCGUGGCUCGCAGCCACCUUU__AAUAAAGAUUUAAGCUGCAAUCAGGA .....(((......((((((....))))))..........................(((((((....))))))).)))..(((((......................)))))........ (-22.21 = -22.40 + 0.19)

| Location | 19,615,836 – 19,615,956 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.36 |
| Shannon entropy | 0.34782 |
| G+C content | 0.53525 |
| Mean single sequence MFE | -37.11 |
| Consensus MFE | -17.93 |
| Energy contribution | -19.60 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.579694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19615836 120 + 21146708 GAGCCACGGUAGCCAUGGCCACCAGCCAGCCAUUGGUGGCGGGAAGCGAGUGCCAGAAAUCUGGCAGAGCUCGGCAGAACAUCAAGCAAUUGUUGUACCCGUUGGUCCGAGUUUAUUAAA (..((((((..(((...((((((((.......))))))))..((.((...((((((....))))))..)))))))...(((.(((....))).)))..))).)))..)............ ( -45.60, z-score = -2.38, R) >droSim1.chr2R 18181919 111 + 19596830 GAGCCACGGUAGCCAUGGCCACCAG--AGCCAU-------GGGAGCCGAGUGCCAGAAAUCUGGCAGAGCUCGGCAGAGCAUCAAGCAAUUGCCGCACCCGUUGGUCCGAGUUUAUUAAA (..((((((...((((((((....)--.)))))-------))..((((((((((((....)))))...)))))))...((..(((....)))..))..))).)))..)............ ( -46.80, z-score = -3.95, R) >droYak2.chr2R 19593764 96 + 21139217 --GCCACAGUAGCCAUGACCACCGG----CCAU------------UGAUGUGCCAGAAAUCCAGCAAAACUCGG---AACAUCAAGCAA--AUUGC-UCCGUAGGUCCGAACUUAUUAAA --..............((((..(((----...(------------((((((.((((.............)).))---.)))))))((..--...))-.)))..))))............. ( -18.92, z-score = -0.27, R) >consensus GAGCCACGGUAGCCAUGGCCACCAG__AGCCAU_______GGGA_CCGAGUGCCAGAAAUCUGGCAGAGCUCGGCAGAACAUCAAGCAAUUGUUGCACCCGUUGGUCCGAGUUUAUUAAA .......(((.((....)).)))...........................((((((....))))))((((((((...(((.....((.......))....)))...))))))))...... (-17.93 = -19.60 + 1.67)

| Location | 19,615,836 – 19,615,956 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.36 |
| Shannon entropy | 0.34782 |
| G+C content | 0.53525 |
| Mean single sequence MFE | -39.70 |
| Consensus MFE | -28.00 |
| Energy contribution | -26.57 |
| Covariance contribution | -1.43 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
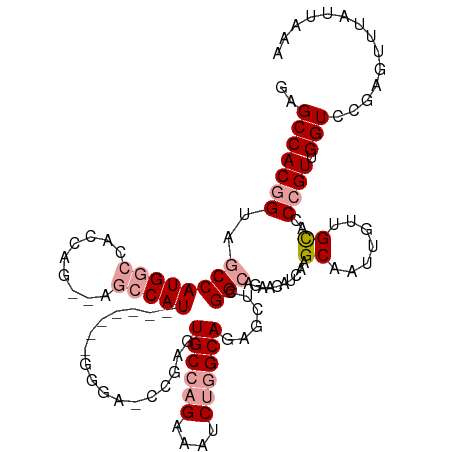
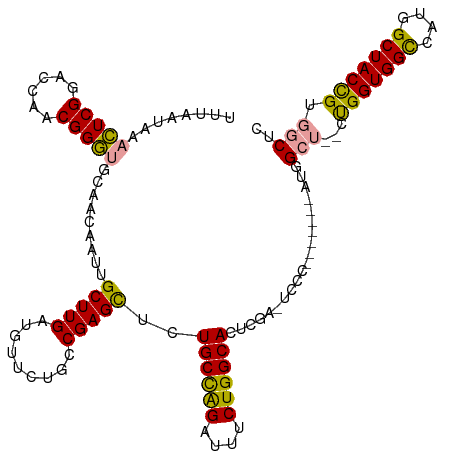
>dm3.chr2R 19615836 120 - 21146708 UUUAAUAAACUCGGACCAACGGGUACAACAAUUGCUUGAUGUUCUGCCGAGCUCUGCCAGAUUUCUGGCACUCGCUUCCCGCCACCAAUGGCUGGCUGGUGGCCAUGGCUACCGUGGCUC .........(((((..((.((((((.......)))))).)).....)))))...((((((....))))))..((((..((((((....)))).))..))))(((((((...))))))).. ( -42.90, z-score = -2.18, R) >droSim1.chr2R 18181919 111 - 19596830 UUUAAUAAACUCGGACCAACGGGUGCGGCAAUUGCUUGAUGCUCUGCCGAGCUCUGCCAGAUUUCUGGCACUCGGCUCCC-------AUGGCU--CUGGUGGCCAUGGCUACCGUGGCUC ........((.(((((((..(((.((((((...((.....))..))))(((...((((((....))))))))).)).)))-------.))).)--)))))((((((((...)))))))). ( -48.00, z-score = -3.57, R) >droYak2.chr2R 19593764 96 - 21139217 UUUAAUAAGUUCGGACCUACGGA-GCAAU--UUGCUUGAUGUU---CCGAGUUUUGCUGGAUUUCUGGCACAUCA------------AUGG----CCGGUGGUCAUGGCUACUGUGGC-- ................(((((((-((((.--..(((((.....---.))))).))))).(((..(((((.((...------------.)))----))))..))).......)))))).-- ( -28.20, z-score = -0.90, R) >consensus UUUAAUAAACUCGGACCAACGGGUGCAACAAUUGCUUGAUGUUCUGCCGAGCUCUGCCAGAUUUCUGGCACUCGA_UCCC_______AUGGCU__CUGGUGGCCAUGGCUACCGUGGCUC ........(((((......))))).........(((((.........)))))..((((((....))))))..........................(((((((....)))))))...... (-28.00 = -26.57 + -1.43)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:46:26 2011