| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,758,900 – 3,759,006 |
| Length | 106 |
| Max. P | 0.500000 |

| Location | 3,758,900 – 3,759,006 |
|---|---|
| Length | 106 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 65.27 |
| Shannon entropy | 0.75322 |
| G+C content | 0.42185 |
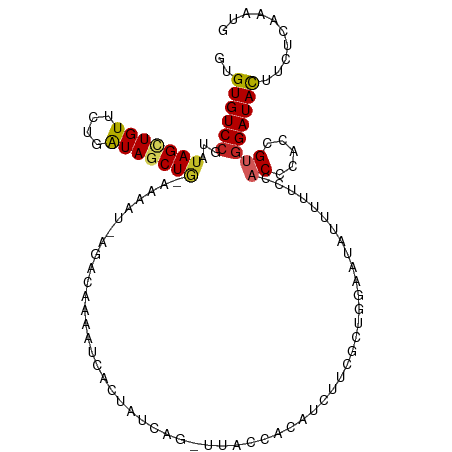
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -8.32 |
| Energy contribution | -8.13 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
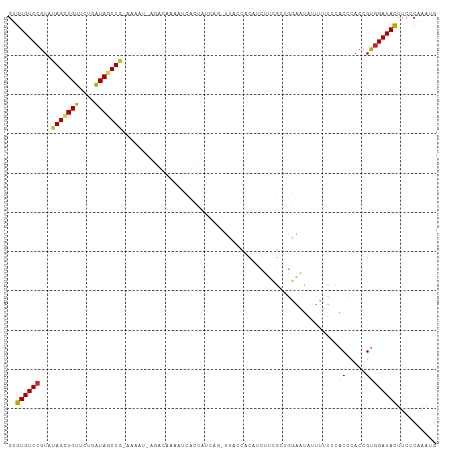
>dm3.chr2L 3758900 106 - 23011544 GUGUGUCCGUAUAGCUGUUCUGGUAGCUGGAAAAU-UGAUAAAAUUGCU-UUAG-UUACCACAUCUUUGCUGAAAUAUUUUUGCACCCACCGUGGAUAUUUUUCAAAUG ..((((((...((((.(...(((((((((((.(((-(.....))))..)-))))-))))))...)...))))..........((.......)))))))).......... ( -22.20, z-score = -1.03, R) >droSim1.chr2L 3714057 106 - 22036055 GUGUGUCCGUGUAGCUGUUCUGGUAGCUGCAAAAU-AGACAAAUUCUCU-UUAG-UUACCACAUCUUUGCUGAAAUAUUUUUCCACCCACCGUGGAUAUUUUUCAAAUG (((((((..((((((((......))))))))....-.))))((((....-..))-))..)))........(((((.((...(((((.....))))).)).))))).... ( -22.30, z-score = -1.76, R) >droSec1.super_5 1854044 106 - 5866729 GUGUGUCCGUGUAGCUGUUCUGGUAGCUGCAAAUU-AGACAAAUUCUCU-UUAG-UUACCACAUCUUUGCUGAAAUAGUUUUCCACCCACCGUGGAUAUUUUUCAAAUG (((((....((((((((......))))))))((((-(((.........)-))))-))..)))))......(((((......(((((.....)))))....))))).... ( -22.30, z-score = -1.41, R) >droYak2.chr2L 3760589 108 - 22324452 GUGUGUCCGUGUAGCUGUUCUGGUAGCUGUCGAAUAAGGCAAAAACACUAUCAG-UUACCAUGACUUCGCUGGAAUGUUUGUCCACCCACCGUGGAUAUUUUUCAAAUG (((.(((...(((((((.....((...((((......))))...)).....)))-))))...)))..)))(((((....(((((((.....)))))))..))))).... ( -29.20, z-score = -1.71, R) >droEre2.scaffold_4929 3804877 107 - 26641161 GUGUGUCCGUGUAGCUGUUCUGGUAGCUGUAAGAU--ACCGAAAUCGAUUUCAGAUUACCAUGCCUGCGCGGGAAUGUCUGUCCACCCACCGUGGAUACUUCUCAAAUG .....(((((((((..((..(((((((((...(((--......))).....))).)))))).)))))))))))......(((((((.....)))))))........... ( -31.00, z-score = -1.70, R) >dp4.chr4_group3 6721907 105 + 11692001 GUGUGUCCGUAUAGUUGUUCUGAUAUCUA-AGGAUACGAUUUGAUUAUUCUC---GCCGCAUAUCUGCAGAUGUAUCGACUGCUACACACCGUGGAUACUUCUCGAAUG ..(((((((((((((((.....((((((.-.(((((.(...(((......))---)...).)))))..))))))..)))))).)))((...)))))))).......... ( -23.30, z-score = 0.08, R) >droPer1.super_1 3820149 105 + 10282868 GUGUGUCCGUAUAGUUGUUCUGAUAUCUA-AAGAUACGAUUUGAUUAUUCUC---GCCGCAUAUCUUCGGAUGUAUCGACUGCUACACACCGUGGAUACUUCUCGAAUG ..(((((((((((((((.....(((((..-((((((.(...(((......))---)...).))))))..)))))..)))))).)))((...)))))))).......... ( -24.90, z-score = -0.90, R) >droMoj3.scaffold_6500 14444180 102 + 32352404 GCGUGUCCGUAUAGCUGUUCUGAUAGCUGCGAGAG-AAACACGAAGUUGAUAAG--GGAGAGAUUUCAUCCACG--ACUCAA--ACUCACCGCGGAUACUUCUCGAAGG ..((((((((.(((((((....)))))))......-......((..((((...(--((((......).))).).--..))))--..))...)))))))).......... ( -25.80, z-score = 0.06, R) >droVir3.scaffold_12963 5254975 101 + 20206255 GUGUGUCCGUAUAGUUGUUUUGAUAGCUAAACAA--AAAUCAAAUUAGCAA----AUACUGCAAAAACAUUUGAUUGCUUGG--ACGCACCGCGGAUACUUCUCGAAGG ((((((((((.(((((((....))))))).))..--.((((((((..(((.----....)))......))))))))....))--))))))((.(((....))))).... ( -27.10, z-score = -2.70, R) >droGri2.scaffold_15126 4428287 104 - 8399593 GUGUGUCAGUGUAGUUGUUUUGAUAGCU--AAACC-AAACGAAAUGUCAACAAGCCAUACAAAUUUUAAUGACGUUACGCAU--ACGCACCGUGGAUACUUCUCGAAGG ((((((..((((((((((....))))))--.....-......(((((((....................)))))))))))..--))))))((.(((....))))).... ( -19.15, z-score = 0.72, R) >consensus GUGUGUCCGUAUAGCUGUUCUGAUAGCUG_AAAAU_AGACAAAAUCACUAUCAG_UUACCACAUCUUCGCUGGAAUAUUUUUCCACCCACCGUGGAUACUUCUCAAAUG ..((((((...(((((((....))))))).......................................................((.....)))))))).......... ( -8.32 = -8.13 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:14:36 2011