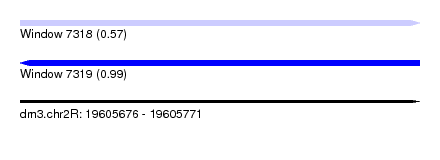
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,605,676 – 19,605,771 |
| Length | 95 |
| Max. P | 0.986741 |

| Location | 19,605,676 – 19,605,771 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 70.94 |
| Shannon entropy | 0.50265 |
| G+C content | 0.48404 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -9.88 |
| Energy contribution | -11.42 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.567164 |
| Prediction | RNA |
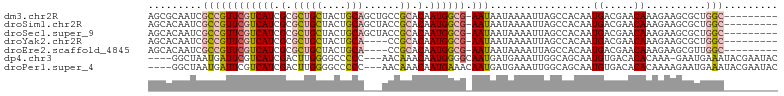
Download alignment: ClustalW | MAF
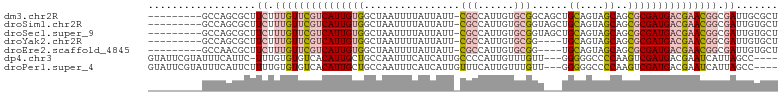
>dm3.chr2R 19605676 95 + 21146708 AGCGCAAUCGCCGUUCGUCAUCGCGCUGCUACUGCAGCUGCCGCACAAUGGCG-AAUAAUAAAAUUAGCCACAAUGACGAACAAAGAAGCGCUGGC--------- (((((..((...(((((((((.(((((((....))))).)).......((((.-(((......))).))))..)))))))))...)).)))))...--------- ( -36.40, z-score = -2.99, R) >droSim1.chr2R 18171979 95 + 19596830 AGCACAAUCGCCGUUCGUCAUCGCGCUGCUACUGCAGCUACCGCACAAUGGCG-AAUAAUAAAAUUAGCCACAAUGACGAACAAAGAAGCGCUGGC--------- (((.(..((...(((((((((.(((((((....)))))....))....((((.-(((......))).))))..)))))))))...)).).)))...--------- ( -29.70, z-score = -2.10, R) >droSec1.super_9 2886443 95 + 3197100 AGCACAAUCGCCGUUCGUCAUCGCGCUGCUACUGCAGCUACCGCACAAUGGCG-AAUAAUAAAAUUAGCCACAAUGACGAACAAAGAAGCGCUGGC--------- (((.(..((...(((((((((.(((((((....)))))....))....((((.-(((......))).))))..)))))))))...)).).)))...--------- ( -29.70, z-score = -2.10, R) >droYak2.chr2R 19583540 91 + 21139217 AGCACAAUCGCCGUUCGUCAUCGCGCUGCUACUGCA----CCGCACAAUGGCG-AAUAAUAAAAUUAGCCACAAUGACGAACAAAGAAGCGCUGGC--------- (((.(..((...(((((((((.(((.(((....)))----.)))....((((.-(((......))).))))..)))))))))...)).).)))...--------- ( -29.10, z-score = -2.49, R) >droEre2.scaffold_4845 20967848 91 + 22589142 AGCACAAUCGCCGUUCGUCAUCGCGCUGCUACUGCA----CCGCACAAUGGCG-AAUAAUAAAAUUAGCCACAAUGACGAACAAAGAAGCGUUGGC--------- .((.(((.(((.(((((((((.(((.(((....)))----.)))....((((.-(((......))).))))..)))))))))......))))))))--------- ( -30.30, z-score = -2.95, R) >dp4.chr3 10384393 97 - 19779522 ----GGCUAAUGAUUCGUCAUCGACUUGGGGCCCCC---AACAAACAAUGGGGCAAUGAUGAAAUUGGCAGCAAUGUGACACACAAA-GAAUGAAAUACGAAUAC ----.((((((..((((((((...(....)(((((.---..........))))).)))))))))))))).....((((...))))..-................. ( -21.90, z-score = -0.85, R) >droPer1.super_4 2445281 98 - 7162766 ----GGCUAAUGAUUCGUCAUCGACUUGGGGCCCCC---AACAAACAAUGAAACAAUGAUGAAAUUGGCAGCAAUGUGACACACAAAAGAAUGAAAUACGAAUAC ----.((((((..((((((((.(..(((((...)))---))((.....))...).)))))))))))))).....((((...)))).................... ( -17.70, z-score = -0.61, R) >consensus AGCACAAUCGCCGUUCGUCAUCGCGCUGCUACUGCA___ACCGCACAAUGGCG_AAUAAUAAAAUUAGCCACAAUGACGAACAAAGAAGCGCUGGC_________ ............(((((((((.(....((....)).............((((...............))))).)))))))))....................... ( -9.88 = -11.42 + 1.53)
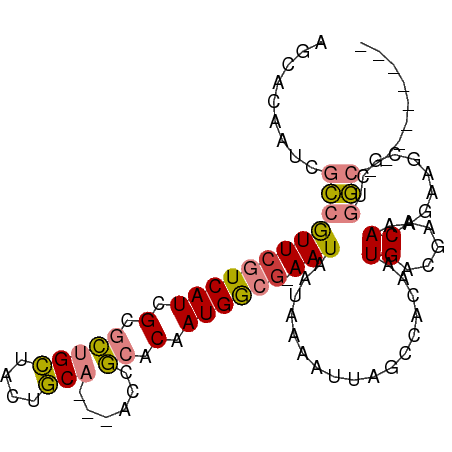
| Location | 19,605,676 – 19,605,771 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 70.94 |
| Shannon entropy | 0.50265 |
| G+C content | 0.48404 |
| Mean single sequence MFE | -34.04 |
| Consensus MFE | -17.30 |
| Energy contribution | -18.53 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19605676 95 - 21146708 ---------GCCAGCGCUUCUUUGUUCGUCAUUGUGGCUAAUUUUAUUAUU-CGCCAUUGUGCGGCAGCUGCAGUAGCAGCGCGAUGACGAACGGCGAUUGCGCU ---------...(((((.((.(((((((((((((((((.(((......)))-.)))...........(((((....))))))))))))))))))).))..))))) ( -42.60, z-score = -3.33, R) >droSim1.chr2R 18171979 95 - 19596830 ---------GCCAGCGCUUCUUUGUUCGUCAUUGUGGCUAAUUUUAUUAUU-CGCCAUUGUGCGGUAGCUGCAGUAGCAGCGCGAUGACGAACGGCGAUUGUGCU ---------...(((((.((.(((((((((((((((((.(((......)))-.)))...........(((((....))))))))))))))))))).))..))))) ( -40.70, z-score = -3.45, R) >droSec1.super_9 2886443 95 - 3197100 ---------GCCAGCGCUUCUUUGUUCGUCAUUGUGGCUAAUUUUAUUAUU-CGCCAUUGUGCGGUAGCUGCAGUAGCAGCGCGAUGACGAACGGCGAUUGUGCU ---------...(((((.((.(((((((((((((((((.(((......)))-.)))...........(((((....))))))))))))))))))).))..))))) ( -40.70, z-score = -3.45, R) >droYak2.chr2R 19583540 91 - 21139217 ---------GCCAGCGCUUCUUUGUUCGUCAUUGUGGCUAAUUUUAUUAUU-CGCCAUUGUGCGG----UGCAGUAGCAGCGCGAUGACGAACGGCGAUUGUGCU ---------...(((((.((.(((((((((((((((((.(((......)))-.)))))...(((.----(((....))).))))))))))))))).))..))))) ( -39.50, z-score = -3.66, R) >droEre2.scaffold_4845 20967848 91 - 22589142 ---------GCCAACGCUUCUUUGUUCGUCAUUGUGGCUAAUUUUAUUAUU-CGCCAUUGUGCGG----UGCAGUAGCAGCGCGAUGACGAACGGCGAUUGUGCU ---------(((((((((.....(((((((((((((((.(((......)))-.)))))...(((.----(((....))).))))))))))))))))).))).)). ( -38.00, z-score = -3.61, R) >dp4.chr3 10384393 97 + 19779522 GUAUUCGUAUUUCAUUC-UUUGUGUGUCACAUUGCUGCCAAUUUCAUCAUUGCCCCAUUGUUUGUU---GGGGGCCCCAAGUCGAUGACGAAUCAUUAGCC---- (.((((((...((....-..((((...))))(((..((((((......))))(((((........)---))))))..)))...))..))))))).......---- ( -18.70, z-score = 0.57, R) >droPer1.super_4 2445281 98 + 7162766 GUAUUCGUAUUUCAUUCUUUUGUGUGUCACAUUGCUGCCAAUUUCAUCAUUGUUUCAUUGUUUGUU---GGGGGCCCCAAGUCGAUGACGAAUCAUUAGCC---- (.((((((...((.......((((...))))..(((.(((((..((............))...)))---)).)))........))..))))))).......---- ( -18.10, z-score = 0.30, R) >consensus _________GCCAGCGCUUCUUUGUUCGUCAUUGUGGCUAAUUUUAUUAUU_CGCCAUUGUGCGGU___UGCAGUAGCAGCGCGAUGACGAACGGCGAUUGUGCU ..................((.(((((((((((((((................(((......)))......((....))..))))))))))))))).))....... (-17.30 = -18.53 + 1.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:46:20 2011