| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,599,582 – 19,599,694 |
| Length | 112 |
| Max. P | 0.988369 |

| Location | 19,599,582 – 19,599,694 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.89 |
| Shannon entropy | 0.29052 |
| G+C content | 0.49727 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -22.42 |
| Energy contribution | -22.68 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
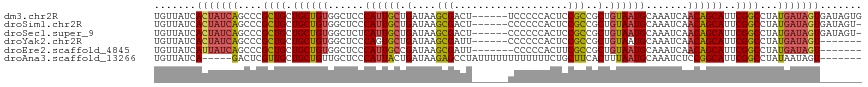
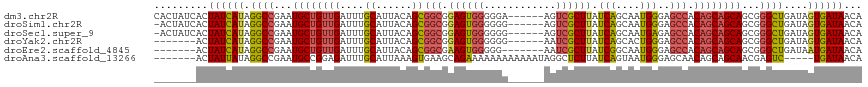
>dm3.chr2R 19599582 112 + 21146708 UGUUAUCACUAUCAGCCCGCUGCUGCUGUGGCUCCCAUUGCUGAUAAGCGACU------UCCCCCACUCCGCCGCUGUAAUGCAAAUCAACAGCAUUCGGCCUAUGAUAGUGAUAGUG ..(((((((((((((((...(((((..((((......(((((....)))))..------....))))......((......)).......)))))...)))...)))))))))))).. ( -36.70, z-score = -3.49, R) >droSim1.chr2R 18165667 111 + 19596830 UGUUAUCACUAUCAGCCCGCUGCUGCUGUGGCUCCCAUUGCUGAUAAGCGACU------CCCCCCACUCCGCCGCUGUAAUGCAAAUCAACAGCAUUCGGCCUAUGAUAGUGAUAGU- ..(((((((((((((((...(((((..((((......(((((....)))))..------....))))......((......)).......)))))...)))...)))))))))))).- ( -36.70, z-score = -3.61, R) >droSec1.super_9 2878575 111 + 3197100 UGUUAUCACUAUCAGCCCGCUGCUGCUGUGGCUCUCAUUGCUGAUAAGCGACU------CCCCCCACUCCGCCGCUGUAAUGCAAAUCAACAGCAUUCGGCCUAUGAUAGUGAUAGU- ..(((((((((((((((...(((((..((((......(((((....)))))..------....))))......((......)).......)))))...)))...)))))))))))).- ( -36.70, z-score = -3.41, R) >droYak2.chr2R 19576780 105 + 21139217 UGUUAUCACUAUCAGCCCGCUGCUGCUGUGGCUCCCAGUGCUGAUAAGCGAUU------CCCCCCACUCCGCCGCUGUAAUGCAAAUCAACAGCAUUCGGCCUAUGAUAGU------- .......(((((((....((((.(((((((((.......(((....)))(...------.....).....)))((......))......))))))..))))...)))))))------- ( -28.60, z-score = -1.40, R) >droEre2.scaffold_4845 20961435 104 + 22589142 UGUUAUCAUUAUCAGCCCGCUGCUGCUGUGGCUCCCAUUGCCGAUAAGCGAUU-------CCCCCACUUCGCCGCUGUAAUGCAAAUCAACAGCAUUCGGCCUAUGAUAGU------- .......(((((((....((((.(((((((((.......)))(((..((((..-------........)))).((......))..))).))))))..))))...)))))))------- ( -29.80, z-score = -2.09, R) >droAna3.scaffold_13266 8237740 106 + 19884421 UGUUAUCA-----GACUCGUUGCUGCUGUUGCUCCCAUUACUGAUAAGAGCCUAUUUUUUUUUUUUCUGCUUCACUUUAAUGCAAAUCUCCGGCAUUCGGCCUAUAAUAGU------- ((((((.(-----(.(.((.(((((...((((.........(((..((((..............))))...))).......)))).....)))))..))).))))))))..------- ( -14.63, z-score = 1.01, R) >consensus UGUUAUCACUAUCAGCCCGCUGCUGCUGUGGCUCCCAUUGCUGAUAAGCGACU______CCCCCCACUCCGCCGCUGUAAUGCAAAUCAACAGCAUUCGGCCUAUGAUAGU_______ .......(((((((....((((.((((((......((((((.(....(((...................)))..).)))))).......))))))..))))...)))))))....... (-22.42 = -22.68 + 0.26)
| Location | 19,599,582 – 19,599,694 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.89 |
| Shannon entropy | 0.29052 |
| G+C content | 0.49727 |
| Mean single sequence MFE | -38.25 |
| Consensus MFE | -28.04 |
| Energy contribution | -28.55 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.988369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
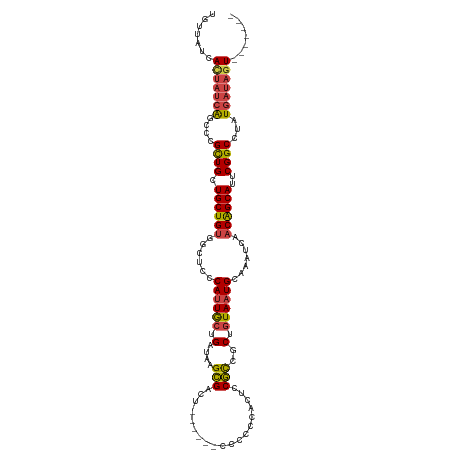
>dm3.chr2R 19599582 112 - 21146708 CACUAUCACUAUCAUAGGCCGAAUGCUGUUGAUUUGCAUUACAGCGGCGGAGUGGGGGA------AGUCGCUUAUCAGCAAUGGGAGCCACAGCAGCAGCGGGCUGAUAGUGAUAACA ...(((((((((((..(.(((..((((((((....((......))(((.((((((....------..)))))).(((....)))..))).)))))))).))).))))))))))))... ( -45.10, z-score = -3.99, R) >droSim1.chr2R 18165667 111 - 19596830 -ACUAUCACUAUCAUAGGCCGAAUGCUGUUGAUUUGCAUUACAGCGGCGGAGUGGGGGG------AGUCGCUUAUCAGCAAUGGGAGCCACAGCAGCAGCGGGCUGAUAGUGAUAACA -..(((((((((((..(.(((..((((((((....((......))(((.((((((....------..)))))).(((....)))..))).)))))))).))).))))))))))))... ( -45.10, z-score = -3.90, R) >droSec1.super_9 2878575 111 - 3197100 -ACUAUCACUAUCAUAGGCCGAAUGCUGUUGAUUUGCAUUACAGCGGCGGAGUGGGGGG------AGUCGCUUAUCAGCAAUGAGAGCCACAGCAGCAGCGGGCUGAUAGUGAUAACA -..(((((((((((..(.(((..((((((((....((......))(((.((((((....------..)))))).(((....)))..))).)))))))).))).))))))))))))... ( -46.00, z-score = -4.27, R) >droYak2.chr2R 19576780 105 - 21139217 -------ACUAUCAUAGGCCGAAUGCUGUUGAUUUGCAUUACAGCGGCGGAGUGGGGGG------AAUCGCUUAUCAGCACUGGGAGCCACAGCAGCAGCGGGCUGAUAGUGAUAACA -------(((((((..(.(((..((((((((....((......))(((.((((((....------..)))))).(((....)))..))).)))))))).))).))))))))....... ( -37.20, z-score = -1.97, R) >droEre2.scaffold_4845 20961435 104 - 22589142 -------ACUAUCAUAGGCCGAAUGCUGUUGAUUUGCAUUACAGCGGCGAAGUGGGGG-------AAUCGCUUAUCGGCAAUGGGAGCCACAGCAGCAGCGGGCUGAUAAUGAUAACA -------..((((((..(((((.((((((((..........))))))))((((((...-------..)))))).)))))..((..((((...((....)).))))..))))))))... ( -34.80, z-score = -1.78, R) >droAna3.scaffold_13266 8237740 106 - 19884421 -------ACUAUUAUAGGCCGAAUGCCGGAGAUUUGCAUUAAAGUGAAGCAGAAAAAAAAAAAAUAGGCUCUUAUCAGUAAUGGGAGCAACAGCAGCAACGAGUC-----UGAUAACA -------.......(((((((..(((.(....(((((.((.....)).)))))..............((((((((.....))))))))..).)))....)).)))-----))...... ( -21.30, z-score = -0.64, R) >consensus _______ACUAUCAUAGGCCGAAUGCUGUUGAUUUGCAUUACAGCGGCGGAGUGGGGGG______AGUCGCUUAUCAGCAAUGGGAGCCACAGCAGCAGCGGGCUGAUAGUGAUAACA .........((((((.((((...((((((((....((......))(((.((((((............)))))).(((....)))..))).))))))))...))))....))))))... (-28.04 = -28.55 + 0.51)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:46:17 2011