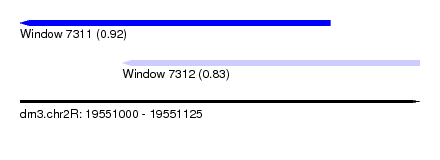
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,551,000 – 19,551,125 |
| Length | 125 |
| Max. P | 0.923535 |

| Location | 19,551,000 – 19,551,097 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 63.12 |
| Shannon entropy | 0.61091 |
| G+C content | 0.42721 |
| Mean single sequence MFE | -31.74 |
| Consensus MFE | -9.48 |
| Energy contribution | -11.20 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19551000 97 - 21146708 -------CCUUUGCACA-GGCACUUUAAUCUAUAUGCAGUGU--GACCUUGCUCUUAAAGUAAGGUC--CAAAUUAUUUUGCGAGCUAAG-GUCACGCUUAUUUUGAGCU------- -------.....((.((-((((..(......)..)))(((((--((((((((((.((((((((....--....)))))))).)))).)))-))))))))....))).)).------- ( -32.70, z-score = -3.97, R) >droSec1.super_9 2830555 89 - 3197100 ---------------CA-GGCCCUUUAAUCUAGGUGCAGUGU--GACCUUGCUCUCAAAGUAAGGUC--CAAAUUAGUUUGCCAGCUAAG-GUCACGCCUAUCUUGAGCU------- ---------------..-.(((((.......))).)).((((--(((((((((..((((.(((....--....))).))))..))).)))-)))))))............------- ( -28.90, z-score = -2.47, R) >droYak2.chr2R 19527282 104 - 21139217 -------CCUUUGCUUA-GGUCCUUUAAGCUGGCUGCAGUGU--GACCUUGCUCUCAAAGUAAGGUC--CAAAUUAUUUUGCGUGCUAUG-GUCACGCUUAGCUUGAGCUGAGGCAU -------((((.(((((-(((....((((((((((..((..(--(((((((((.....)))))))))--((((....)))).)..))..)-)))).))))))))))))).))))... ( -40.90, z-score = -3.47, R) >droEre2.scaffold_4845 20913025 104 - 22589142 -------CCCUUGCAUA-GGCCCUUUAGGCUAGCUGCAGUGU--GACCUUGCUCUUAAAGUAGGGUC--CAAAUUAUUUCGCGAGCUAAG-GUCACGCUUAGCUUGAGCUAAGGCAC -------.((((((...-((((.....))))(((((.(((((--((((((((((...((((((....--....))))))...)))).)))-)))))))))))))...)).))))... ( -41.40, z-score = -2.83, R) >droAna3.scaffold_13266 8184331 110 - 19884421 CCAUUGCCUCUAUCGUAUAACAAUAAAAUCUAAAUUUGUUUUUAAGAACUGACCUUUAGGUAUAAUCGGGAAAUUGGUAAGUAAAUUAAGUACUACCUCUUUAUUUACCA------- ...((.((.........(((.((((((.......)))))).))).((....(((....)))....)))).))..(((((((((((...((......)).)))))))))))------- ( -14.80, z-score = 0.43, R) >consensus _______CCCUUGCAUA_GGCCCUUUAAUCUAGAUGCAGUGU__GACCUUGCUCUUAAAGUAAGGUC__CAAAUUAUUUUGCGAGCUAAG_GUCACGCUUAUCUUGAGCU_______ ...............................(((((.(((((..((((((((((.((((((((..........)))))))).)))).))).)))))))))))))............. ( -9.48 = -11.20 + 1.72)


| Location | 19,551,032 – 19,551,125 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 54.97 |
| Shannon entropy | 0.73007 |
| G+C content | 0.40394 |
| Mean single sequence MFE | -19.98 |
| Consensus MFE | -8.06 |
| Energy contribution | -8.14 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.825836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19551032 93 - 21146708 -------------------GCUAACUUUUGCCAUGCCAAACUCCAAUCCUUUGCACA-GGCACUUUAAUCUAUAUGCAGUGU--GACCUUGCUCUUAAAGUAAGGUC--CAAAUUAU -------------------.........((((.(((.(((.........))))))..-)))).................((.--(((((((((.....)))))))))--))...... ( -18.70, z-score = -1.28, R) >droSec1.super_9 2830587 74 - 3197100 -------------------------------------UGAUUUC-AUGCCAAACUCA-GGCCCUUUAAUCUAGGUGCAGUGU--GACCUUGCUCUCAAAGUAAGGUC--CAAAUUAG -------------------------------------.((((..-..(((.......-))).....)))).........((.--(((((((((.....)))))))))--))...... ( -18.20, z-score = -1.64, R) >droYak2.chr2R 19527321 93 - 21139217 -------------------GCUUUUUUCGUCCAAGCCAAACUCCAAUCCUUUGCUUA-GGUCCUUUAAGCUGGCUGCAGUGU--GACCUUGCUCUCAAAGUAAGGUC--CAAAUUAU -------------------..............(((((.........(((......)-))..........)))))....((.--(((((((((.....)))))))))--))...... ( -21.71, z-score = -1.52, R) >droEre2.scaffold_4845 20913064 93 - 22589142 -------------------GCUUUUUUUAUCCAUGCCAAACUCCAAUCCCUUGCAUA-GGCCCUUUAGGCUAGCUGCAGUGU--GACCUUGCUCUUAAAGUAGGGUC--CAAAUUAU -------------------...............................(((((..-((((.....))))...)))))((.--(((((((((.....)))))))))--))...... ( -21.80, z-score = -0.64, R) >droAna3.scaffold_13266 8184364 117 - 19884421 GUUUGAUAUAAAUAAGUCCAAUUCACCCUGAUAUGAUAAGCCAUUGCCUCUAUCGUAUAACAAUAAAAUCUAAAUUUGUUUUUAAGAACUGACCUUUAGGUAUAAUCGGGAAAUUGG .................((((((..(((..((((((((.((....))...)))))))).........(((((((...((((....)))).....)))))))......))).)))))) ( -19.50, z-score = -0.68, R) >consensus ___________________GCUUUUUUCUGCCAUGCCAAACUCCAAUCCCUUGCAUA_GGCCCUUUAAUCUAGAUGCAGUGU__GACCUUGCUCUUAAAGUAAGGUC__CAAAUUAU ...............................................................................((...(((((((((.....)))))))))..))...... ( -8.06 = -8.14 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:46:14 2011