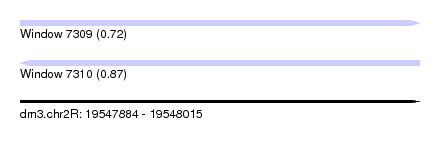
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,547,884 – 19,548,015 |
| Length | 131 |
| Max. P | 0.872942 |

| Location | 19,547,884 – 19,548,015 |
|---|---|
| Length | 131 |
| Sequences | 4 |
| Columns | 131 |
| Reading direction | forward |
| Mean pairwise identity | 80.60 |
| Shannon entropy | 0.29101 |
| G+C content | 0.38629 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -21.10 |
| Energy contribution | -22.73 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.718051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
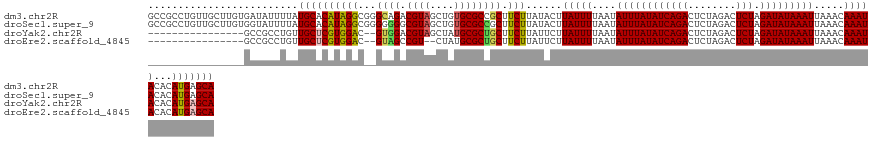
>dm3.chr2R 19547884 131 + 21146708 UGCUCAUGUGUAUUUGUUUAAUUUAUAUCUAGAGUCUAGAGUCUGAUAUAAAUAUUAAAAUAAGUAUAAGAAGCGGCGCACAGCUACGUCUGCCCGCCUAUGUGCAUAAAAUAUCACAAGCAACAGGCGGC .(((....(((((((((((.(((((((((.(((.(....).))))))))))))....)))))))))))...)))((((.((......)).))))(((((...(((..............)))..))))).. ( -35.34, z-score = -2.00, R) >droSec1.super_9 2827329 131 + 3197100 UGCUCAUGUGUAUUUGUUUAAUUUAUAUCUAGAGUCUAGAGUCUGAUAUAAAUAUUAAAAUAAGUAUAAGAAGCGGCGCACAGCUACGCCCCCCCGCCUAUGUGCAUAAAAUACCACAAGCAACAGGCGGC .(((....(((((((((((.(((((((((.(((.(....).))))))))))))....)))))))))))...)))((((........))))...((((((...(((..............)))..)))))). ( -37.64, z-score = -3.26, R) >droYak2.chr2R 19524163 113 + 21139217 UGCUCAUGUGUAUUUGUUUAAUUUAUAUCUAGAGUCUAGAGUCUGAUAUAAAUAUUAAAAUAAGAAUAAGAAGCAGCGCAUAGCUACGUCCAC--GUCCACGAGCAACAGGCGGC---------------- (((((.(((.(((((.....(((((((((.(((.(....).))))))))))))....)))))...))).).)))).(((...(((.(((....--....)))))).....)))..---------------- ( -24.10, z-score = -0.52, R) >droEre2.scaffold_4845 20910042 111 + 22589142 UGCUCAUGUGUAUUUGUUUAAUUUAUAUCUAGAGUCUAGAGUCUGAUAUAAAUAUUAAAAUAAGAAUAAGAAGCAGCGCAUAG--ACGGCUAC--GUCCACGAGCAACAGGCGGC---------------- (((((((((((...(((((..((((((((.(((.(....).)))))))))))((((........))))..))))))))))).(--(((....)--)))...))))).........---------------- ( -27.60, z-score = -1.65, R) >consensus UGCUCAUGUGUAUUUGUUUAAUUUAUAUCUAGAGUCUAGAGUCUGAUAUAAAUAUUAAAAUAAGAAUAAGAAGCAGCGCACAGCUACGUCCAC__GCCCACGAGCAAAAAACAGC________________ (((((((((((((((((((.(((((((((.(((.(....).))))))))))))....)))))))))))....(.(((.....))).)...........))))))))......................... (-21.10 = -22.73 + 1.62)


| Location | 19,547,884 – 19,548,015 |
|---|---|
| Length | 131 |
| Sequences | 4 |
| Columns | 131 |
| Reading direction | reverse |
| Mean pairwise identity | 80.60 |
| Shannon entropy | 0.29101 |
| G+C content | 0.38629 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -20.40 |
| Energy contribution | -21.65 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19547884 131 - 21146708 GCCGCCUGUUGCUUGUGAUAUUUUAUGCACAUAGGCGGGCAGACGUAGCUGUGCGCCGCUUCUUAUACUUAUUUUAAUAUUUAUAUCAGACUCUAGACUCUAGAUAUAAAUUAAACAAAUACACAUGAGCA .(((((((((((..((((....))))))).))))))))(((((.(..((.....))..).)))......(((((....((((((((((((........))).))))))))).....))))).......)). ( -32.00, z-score = -1.41, R) >droSec1.super_9 2827329 131 - 3197100 GCCGCCUGUUGCUUGUGGUAUUUUAUGCACAUAGGCGGGGGGGCGUAGCUGUGCGCCGCUUCUUAUACUUAUUUUAAUAUUUAUAUCAGACUCUAGACUCUAGAUAUAAAUUAAACAAAUACACAUGAGCA .(((((((((((.((........)).))).))))))))((.((((((....)))))).)).(((((...(((((....((((((((((((........))).))))))))).....)))))...))))).. ( -38.10, z-score = -2.15, R) >droYak2.chr2R 19524163 113 - 21139217 ----------------GCCGCCUGUUGCUCGUGGAC--GUGGACGUAGCUAUGCGCUGCUUCUUAUUCUUAUUUUAAUAUUUAUAUCAGACUCUAGACUCUAGAUAUAAAUUAAACAAAUACACAUGAGCA ----------------.........((((((((((.--..(((.(((((.....))))).)))...)).(((((....((((((((((((........))).))))))))).....)))))..)))))))) ( -29.20, z-score = -2.63, R) >droEre2.scaffold_4845 20910042 111 - 22589142 ----------------GCCGCCUGUUGCUCGUGGAC--GUAGCCGU--CUAUGCGCUGCUUCUUAUUCUUAUUUUAAUAUUUAUAUCAGACUCUAGACUCUAGAUAUAAAUUAAACAAAUACACAUGAGCA ----------------.........((((((((((.--(((((((.--...)).))))).)).......(((((....((((((((((((........))).))))))))).....)))))..)))))))) ( -27.20, z-score = -2.76, R) >consensus ________________GCCACCUGAUGCACAUAGAC__GUAGACGUAGCUAUGCGCCGCUUCUUAUACUUAUUUUAAUAUUUAUAUCAGACUCUAGACUCUAGAUAUAAAUUAAACAAAUACACAUGAGCA .........................((((((((((((.((((......)))).))))............(((((....((((((((((((........))).))))))))).....)))))..)))))))) (-20.40 = -21.65 + 1.25)
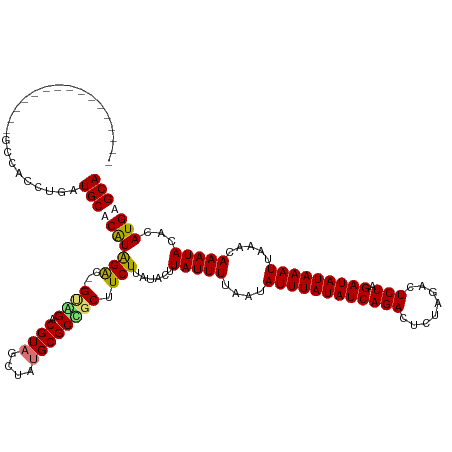

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:46:12 2011