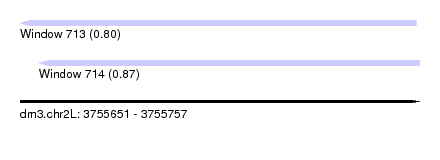
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,755,651 – 3,755,757 |
| Length | 106 |
| Max. P | 0.869921 |

| Location | 3,755,651 – 3,755,756 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 92.64 |
| Shannon entropy | 0.13885 |
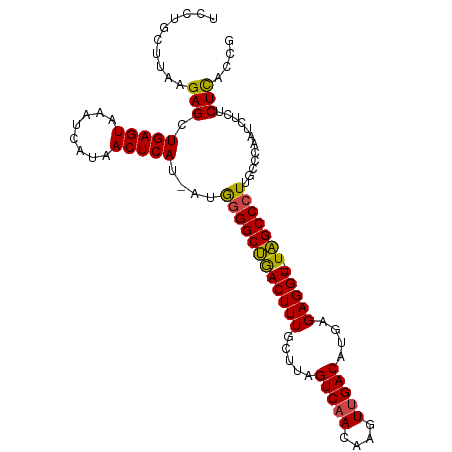
| G+C content | 0.45512 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -23.59 |
| Energy contribution | -23.23 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.798937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
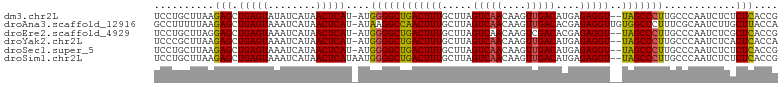
>dm3.chr2L 3755651 105 - 23011544 CCUGCUUAAGAGCUGAGUAUAUCAUAACUCAU-AUGGGGCUGACUUUGCUUAGUCAACAAGUUGACAUGAGAGGU--UAGCCCUUGCCCAAUCUCUCUCACCGCAUCU ..(((...((((.(((((........))))).-..((((((((((...(((((((((....))))).)))).)))--)))))))..........))))....)))... ( -33.20, z-score = -2.58, R) >droAna3.scaffold_12916 5824786 107 - 16180835 CCUUUUUAAGAGCUGAGUAAAUCAUAACUCAU-AUAAGGCCAACUUUGCUUAGUCAACAAGUUGACACGAGAGGUUGUGGCCCUUUCGCAAUCUUGCUUACCACAUCU .........(((((((((........))))).-....((((((((((.....(((((....)))))....)))))..))))).............))))......... ( -24.30, z-score = -1.18, R) >droEre2.scaffold_4929 3801483 105 - 26641161 CCUGCUUAGGAGCUGAGUAAAUCAUAACUCAU-AUGGGGCUGACUUUGCUUAGUCAACAAGUCGACACGAGAGGU--UAGCCCUUGCCCAAUCUCGCUCACCGCAUCU ..(((....(((((((((........))))).-..((((((((((((.....(((........)))....)))))--)))))))...........))))...)))... ( -33.30, z-score = -2.22, R) >droYak2.chr2L 3757169 105 - 22324452 CCCGCUUAAGAGCUGAGUAAAUCAUAACUCAU-AUGGGGCUGACUUUGCUUAGUCAACAAGUUGACAUGAGAGGU--UAGCCCUUGCCCAAUCUCACUCACCACAUCU .........(((.((((.....((((.....)-)))(((((((((...(((((((((....))))).)))).)))--)))))).........)))))))......... ( -31.90, z-score = -2.81, R) >droSec1.super_5 1850817 105 - 5866729 CCUGCUUAAGAGCUGAGUAAAUCAUAACUCAU-AUGGGGCUGACUUUGCUUAGUCAACAAGUUGACAUGAGAGGU--UAGCCCUUGCCCAAUCUCUCUCACCGCAUCU ..(((...((((.(((((........))))).-..((((((((((...(((((((((....))))).)))).)))--)))))))..........))))....)))... ( -33.20, z-score = -2.67, R) >droSim1.chr2L 3710804 106 - 22036055 CCUGCUUAAGAGCUGAGUAAAUCAUAACUCAUAAUGGGGCUGACUUUGCUUAGUCAACAAGUUGACAUGAGAGGU--UAGCCCUUGCCCAAUCUCUCUCACCGCAUCU ..(((...((((.(((((........)))))....((((((((((...(((((((((....))))).)))).)))--)))))))..........))))....)))... ( -33.20, z-score = -2.70, R) >consensus CCUGCUUAAGAGCUGAGUAAAUCAUAACUCAU_AUGGGGCUGACUUUGCUUAGUCAACAAGUUGACAUGAGAGGU__UAGCCCUUGCCCAAUCUCUCUCACCGCAUCU .........(((.(((((........)))))....((((((((((((.....(((((....)))))....)))))..)))))))............)))......... (-23.59 = -23.23 + -0.36)

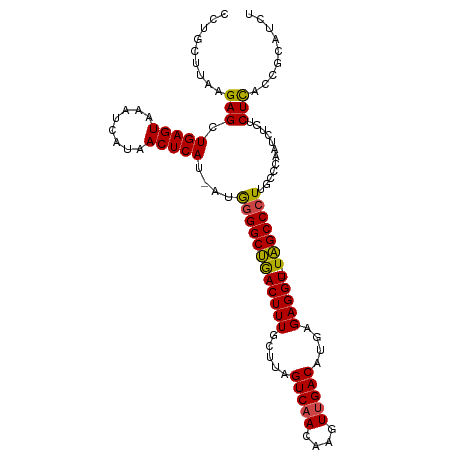
| Location | 3,755,656 – 3,755,757 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 92.03 |
| Shannon entropy | 0.15044 |
| G+C content | 0.45497 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -23.59 |
| Energy contribution | -23.23 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.869921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3755656 101 - 23011544 UCCUGCUUAAGAGCUGAGUAUAUCAUAACUCAU-AUGGGGCUGACUUUGCUUAGUCAACAAGUUGACAUGAGAGGU--UAGCCCUUGCCCAAUCUCUCUCACCG .........((((.(((((........))))).-..((((((((((...(((((((((....))))).)))).)))--)))))))........))))....... ( -30.80, z-score = -2.36, R) >droAna3.scaffold_12916 5824791 103 - 16180835 GCCUUUUUAAGAGCUGAGUAAAUCAUAACUCAU-AUAAGGCCAACUUUGCUUAGUCAACAAGUUGACACGAGAGGUUGUGGCCCUUUCGCAAUCUUGCUUACCA (((((..((.(((.(((.....)))...))).)-).)))))(((..((((...(((((....)))))..((((((.......))))))))))..)))....... ( -26.90, z-score = -1.83, R) >droEre2.scaffold_4929 3801488 101 - 26641161 UCCUGCUUAGGAGCUGAGUAAAUCAUAACUCAU-AUGGGGCUGACUUUGCUUAGUCAACAAGUCGACACGAGAGGU--UAGCCCUUGCCCAAUCUCGCUCACCG ..........(((((((((........))))).-..((((((((((((.....(((........)))....)))))--)))))))...........)))).... ( -30.90, z-score = -1.95, R) >droYak2.chr2L 3757174 101 - 22324452 UCCCGCUUAAGAGCUGAGUAAAUCAUAACUCAU-AUGGGGCUGACUUUGCUUAGUCAACAAGUUGACAUGAGAGGU--UAGCCCUUGCCCAAUCUCACUCACCA ..........(((.((((.....((((.....)-)))(((((((((...(((((((((....))))).)))).)))--)))))).........))))))).... ( -31.90, z-score = -2.91, R) >droSec1.super_5 1850822 101 - 5866729 UCCUGCUUAAGAGCUGAGUAAAUCAUAACUCAU-AUGGGGCUGACUUUGCUUAGUCAACAAGUUGACAUGAGAGGU--UAGCCCUUGCCCAAUCUCUCUCACCG .........((((.(((((........))))).-..((((((((((...(((((((((....))))).)))).)))--)))))))........))))....... ( -30.80, z-score = -2.45, R) >droSim1.chr2L 3710809 102 - 22036055 UCCUGCUUAAGAGCUGAGUAAAUCAUAACUCAUAAUGGGGCUGACUUUGCUUAGUCAACAAGUUGACAUGAGAGGU--UAGCCCUUGCCCAAUCUCUCUCACCG .........((((.(((((........)))))....((((((((((...(((((((((....))))).)))).)))--)))))))........))))....... ( -30.80, z-score = -2.47, R) >consensus UCCUGCUUAAGAGCUGAGUAAAUCAUAACUCAU_AUGGGGCUGACUUUGCUUAGUCAACAAGUUGACAUGAGAGGU__UAGCCCUUGCCCAAUCUCUCUCACCG ..........(((.(((((........)))))....((((((((((((.....(((((....)))))....)))))..)))))))............))).... (-23.59 = -23.23 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:14:36 2011