
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,524,235 – 19,524,359 |
| Length | 124 |
| Max. P | 0.851502 |

| Location | 19,524,235 – 19,524,359 |
|---|---|
| Length | 124 |
| Sequences | 6 |
| Columns | 138 |
| Reading direction | forward |
| Mean pairwise identity | 74.96 |
| Shannon entropy | 0.46166 |
| G+C content | 0.46283 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -18.49 |
| Energy contribution | -20.02 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.851502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
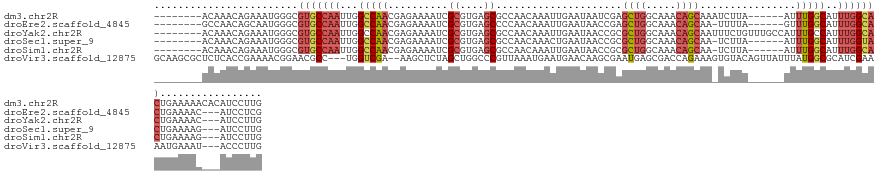
>dm3.chr2R 19524235 124 + 21146708 --------ACAAACAGAAAUGGGCGUGCCAAUUGGCCAACGAGAAAAUCGCGUGAGCGCCAACAAAUUGAAUAAUCGAGCUGGCAAACAGCAAAUCUUA------AUUUGGCAUUUGGCACUGAAAAACACAUCCUUG --------.....((....))...(((((((...(((((.((((....(((....)))....................((((.....))))...)))).------..)))))..)))))))................. ( -35.30, z-score = -2.39, R) >droEre2.scaffold_4845 20886768 120 + 22589142 --------GCCAACAGCAAUGGGCGUGCCAAUUGGCCAACGAGAAAAUCGCGUGAGCCCCAACAAAUUGAAUAACCGAGCUGGCAAACAGCAA-UUUUA------GUUUGGCAUUUGGCACUGAAAAC---AUCCUCG --------(((((..((..((((((((((....)))..)))........((....)))))).(((((((((.......((((.....))))..-.))))------)))))))..))))).........---....... ( -35.40, z-score = -1.55, R) >droYak2.chr2R 19500016 127 + 21139217 --------ACAAACAGAAAUGGGCGUGCCAAUUGGCCAACGAGAAAAUCGCGUGAGCGCCAACAAAUUGAAUAACCGCGCUGGCAAACAGCAAUUUCUGUUUGCCAUUUGCCAUUUGGCACUGAAAAC---AUCCUUG --------.....((....))...(((((((.((((..(((.(.....).))).(((((.................)))))(((((((((......)))))))))....)))).))))))).......---....... ( -41.53, z-score = -2.75, R) >droSec1.super_9 2803847 120 + 3197100 --------ACAAACAGAAAUGGGCGUGCCAAUUGGCCAACGAGAAAAUCGCGUGAGCGCCAACAAACUGAAUAACCGCGCUGGCAAACAGCAA-UCUUA------AUUUGGCAUUUGGUACUGAAAAG---AUCCUUG --------.....((....))...(((((((...(((((.((((....(((....)))....................((((.....))))..-)))).------..)))))..))))))).......---....... ( -33.40, z-score = -1.59, R) >droSim1.chr2R 18094751 120 + 19596830 --------ACAAACAGAAAUGGGCGUGCCAAUUGGCCAACGAGAAAAUCGCGUGAGCGCCAACAAAUUGAAUAACCGCGCUGGCAAACAGCAA-UCUUA------AUUUGGCAUUUGGCACUGAAAAG---AUCCUUG --------.....((....))...(((((((...(((((.((((....(((....)))....................((((.....))))..-)))).------..)))))..))))))).......---....... ( -35.40, z-score = -1.93, R) >droVir3.scaffold_12875 2127511 130 - 20611582 GCAAGCGCUCUCACCGAAAACGGAACGCC---UGGUCGA--AAGCUCUAGCUGGCCCGUUAAAUGAAUGAACAAGCGAAUGAGCGACCAGAAAGUGUACAGUUAUUUAUGGCGCAUCGAAAAUGAAAU---ACCCUUG ....(((((....(((....))).(((((---(((((..--..((((..(((....((((.....))))....)))....))))))))))...))))............)))))..............---....... ( -32.10, z-score = -0.70, R) >consensus ________ACAAACAGAAAUGGGCGUGCCAAUUGGCCAACGAGAAAAUCGCGUGAGCGCCAACAAAUUGAAUAACCGAGCUGGCAAACAGCAA_UCUUA______AUUUGGCAUUUGGCACUGAAAAC___AUCCUUG ........................(((((((...(((((..........(((....)))...................((((.....))))................)))))..)))))))................. (-18.49 = -20.02 + 1.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:46:09 2011