
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,519,714 – 19,519,898 |
| Length | 184 |
| Max. P | 0.888908 |

| Location | 19,519,714 – 19,519,788 |
|---|---|
| Length | 74 |
| Sequences | 5 |
| Columns | 74 |
| Reading direction | forward |
| Mean pairwise identity | 77.52 |
| Shannon entropy | 0.38969 |
| G+C content | 0.46678 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -13.37 |
| Energy contribution | -14.05 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.888908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19519714 74 + 21146708 GCGUUAACAAACGUGCUAGGCAGUGUAUCUGCGUUUUCUGCCCACAUUCUGCGGAUACAGAUACACGAGCACAC ............(((((.....(((((((((.....(((((.........)))))..))))))))).))))).. ( -24.80, z-score = -2.76, R) >droSim1.chr2R 18090327 74 + 19596830 GCGUGAACAAACGUGUUAGGCAGUGUAUCUGCGUUUUCUGCCCACAUUCUGCGGAUACAGAUACACGAGCACAC ((((......)))).....((.(((((((((.....(((((.........)))))..)))))))))..)).... ( -22.90, z-score = -1.71, R) >droSec1.super_9 2799412 74 + 3197100 GCGUGAACAAACGUGUUAGGCAGUGUAUCUGCGUUUUCUGCCCACAUUUUGCGGAUACAGAUACACGAGCACAC ((((......)))).....((.(((((((((.....(((((.........)))))..)))))))))..)).... ( -22.90, z-score = -1.95, R) >droYak2.chr2R 19495236 68 + 21139217 CAGUAAGCUAAUGUGUUAAACAGUGUAUCUGCGUUUUCUGUCCACAUUCUGCGGAUAC------ACGAGCACAC ...........((((((.....(((((((((((.....((....))...)))))))))------)).)))))). ( -20.60, z-score = -2.42, R) >droAna3.scaffold_13266 8151576 64 + 19884421 CAGUUACCUGUAGUGUUAAAUAGUGUAUCUAC--UCUCUG--CGUAUCUGACGGAUAC------ACAAACACAC (((....)))..(((((.....((((((((..--((....--.......)).))))))------)).))))).. ( -12.30, z-score = -0.17, R) >consensus GCGUGAACAAACGUGUUAGGCAGUGUAUCUGCGUUUUCUGCCCACAUUCUGCGGAUACAGAUACACGAGCACAC ............(((((......((((((((((................))))))))))........))))).. (-13.37 = -14.05 + 0.68)



| Location | 19,519,714 – 19,519,788 |
|---|---|
| Length | 74 |
| Sequences | 5 |
| Columns | 74 |
| Reading direction | reverse |
| Mean pairwise identity | 77.52 |
| Shannon entropy | 0.38969 |
| G+C content | 0.46678 |
| Mean single sequence MFE | -21.78 |
| Consensus MFE | -13.12 |
| Energy contribution | -14.44 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.756301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19519714 74 - 21146708 GUGUGCUCGUGUAUCUGUAUCCGCAGAAUGUGGGCAGAAAACGCAGAUACACUGCCUAGCACGUUUGUUAACGC ..(((((.((((((((((.(((((.....)))))..(....))))))))))).....)))))(((....))).. ( -23.60, z-score = -1.28, R) >droSim1.chr2R 18090327 74 - 19596830 GUGUGCUCGUGUAUCUGUAUCCGCAGAAUGUGGGCAGAAAACGCAGAUACACUGCCUAACACGUUUGUUCACGC (((.((.(((((.(((((..((((.....)))))))))....((((.....))))...)))))...)).))).. ( -24.90, z-score = -1.96, R) >droSec1.super_9 2799412 74 - 3197100 GUGUGCUCGUGUAUCUGUAUCCGCAAAAUGUGGGCAGAAAACGCAGAUACACUGCCUAACACGUUUGUUCACGC (((.((.(((((.(((((..((((.....)))))))))....((((.....))))...)))))...)).))).. ( -24.90, z-score = -2.44, R) >droYak2.chr2R 19495236 68 - 21139217 GUGUGCUCGU------GUAUCCGCAGAAUGUGGACAGAAAACGCAGAUACACUGUUUAACACAUUAGCUUACUG (((.(((.((------((.(((((.....)))))........((((.....)))).....)))).))).))).. ( -16.90, z-score = -0.89, R) >droAna3.scaffold_13266 8151576 64 - 19884421 GUGUGUUUGU------GUAUCCGUCAGAUACG--CAGAGA--GUAGAUACACUAUUUAACACUACAGGUAACUG ((((.(((((------(((((.....))))))--))))((--((((.....)))))).))))..(((....))) ( -18.60, z-score = -2.02, R) >consensus GUGUGCUCGUGUAUCUGUAUCCGCAGAAUGUGGGCAGAAAACGCAGAUACACUGCCUAACACGUUUGUUAACGC ((((((..((((((((((.(((((.....)))))........)))))))))).))...))))............ (-13.12 = -14.44 + 1.32)

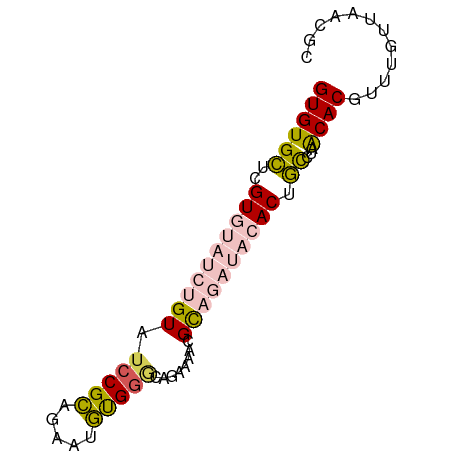

| Location | 19,519,788 – 19,519,898 |
|---|---|
| Length | 110 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.90 |
| Shannon entropy | 0.46984 |
| G+C content | 0.52141 |
| Mean single sequence MFE | -34.56 |
| Consensus MFE | -12.54 |
| Energy contribution | -13.66 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.646213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19519788 110 - 21146708 GCAAUCUAUGCAAUGCCCGAUUCGUG----UGUGUGUGAGAGUGUGCGCAAUACGCCGCCUUCUUAUUCGCUCAUUCCGAUACACUCACACA----CACACGAACACCUGAGGCAGAG (((.....)))..((((((.((((((----((((((((((.(((.(((.....))))))........(((.......)))....))))))))----))))))))....)).))))... ( -41.80, z-score = -4.43, R) >droSim1.chr2R 18090401 108 - 19596830 GCGAUCUAUGCAAUGCCCGAUUCGUG----UGUGUGUGAGAGUGUGCGCAAUACGCCGCCUUCUUAUUCGCUCGUUCCGAUACACUCACACA----CACGC--ACACCUGAGGCAGAG (((.....)))..((((((....(((----((((((((.(((((((((....(((..((..........)).)))..)).))))))).))))----)))))--))...)).))))... ( -38.60, z-score = -2.51, R) >droSec1.super_9 2799486 108 - 3197100 GCAAUCUAUGCAAUGCCCGAUUCGUG----UGUGUGUGAGAGUGUGCGCAAUACGCCGCCUUCUUAUUCGCUCGUUCCGAUACACUCACACA----CACGC--ACACCUGAGGCAGAG (((.....)))..((((((....(((----((((((((.(((((((((....(((..((..........)).)))..)).))))))).))))----)))))--))...)).))))... ( -39.80, z-score = -3.22, R) >droYak2.chr2R 19495304 108 - 21139217 GCAAUCUAUGCACUGCUCGAUUCGUG----UGUGUGUGAGAGUGUCCGCAAUACGCCGCCUUCUUAUUCGCUCGUUCCGAUACACUCACACU----CGCGC--ACACCUGAGGCAGAG (((.....))).(((((((....(((----((.(((((((.(((((.(....(((..((..........)).))).).))))).))))))).----)))))--.....))).)))).. ( -33.90, z-score = -1.32, R) >droEre2.scaffold_4845 20882170 114 - 22589142 GCAAUCUAUGCAAUGCCCGAUUCGUC----UGUGUGUGAGAGUGUCCGCAAUACGCCGCCUUCUUAUUCGCUCGUUCCGAUACACUCACACAGAGGCAAACGCACACCUGAAACAGAG ((((((...((...))..)))).(((----(.((((((((.(((((.(....(((..((..........)).))).).))))).)))))))).))))....))............... ( -29.00, z-score = -0.78, R) >droAna3.scaffold_13266 8151641 110 - 19884421 GCAAUCUAUGCAAUGCCCGAUUCGUGUAUAUGUUUGUGUGAGUAUCUGCUAUACGCCGGCGUCUCAUUCGCCCGUUUCGAUACACACACACG----UGUGCG-AGAGCGGCAAAA--- (((.....)))..((((.(.(((.(((((((((..(((((.(((((.((.....)).((((.......))))......))))).))))))))----))))))-.))))))))...--- ( -37.40, z-score = -1.92, R) >droGri2.scaffold_15245 13720886 97 + 18325388 AUAAUCUAUGCAAUACUAAAGUCGUG----UGUGUAU----------ACAAUGCGCUCGCCGCUCAUUCGUUCGCCAC-ACACACACACACCG-CUACUGCUCGCACUCACAU----- ........(((.........((.(((----(((((..----------((((((.((.....)).)))).)).....))-)))))))).....(-(....))..))).......----- ( -21.40, z-score = -1.57, R) >consensus GCAAUCUAUGCAAUGCCCGAUUCGUG____UGUGUGUGAGAGUGUCCGCAAUACGCCGCCUUCUUAUUCGCUCGUUCCGAUACACUCACACA____CACGC__ACACCUGAGGCAGAG (((.....)))............(((......((((((((.((((.((.(((..((.............))..))).)))))).))))))))......)))................. (-12.54 = -13.66 + 1.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:46:07 2011