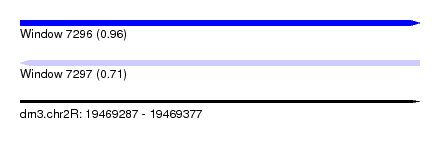
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,469,287 – 19,469,377 |
| Length | 90 |
| Max. P | 0.959354 |

| Location | 19,469,287 – 19,469,377 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 75.18 |
| Shannon entropy | 0.38049 |
| G+C content | 0.46169 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -12.28 |
| Energy contribution | -14.24 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19469287 90 + 21146708 UGACUUGACUGACACUCCUUAGUCUUAGUCAUUAGCCAACUAACUGGCA------------AAUGGCUUGGUAUCUUCGCCCGUCAUUUGUGUGGGCUAGAU---- (((((.((((((......))))))..)))))((((((.....((..(((------------((((((..(((......))).))))))))))).))))))..---- ( -30.50, z-score = -3.23, R) >droSim1.chr2R 18040539 77 + 19596830 UGACUUGACUGACACUCCUUAGUCUUAGUCAUUAGCCAACUAACUGGCA------------AAUGGCGUGGUAUCUUCGCCCGCUAGAU----------------- (((((.((((((......))))))..)))))...((((......)))).------------..(((((.(((......))))))))...----------------- ( -26.20, z-score = -3.78, R) >droSec1.super_9 2750511 77 + 3197100 UGACUUGACUGACACUCCUUAGUCUUAGUCAUUAGCCAACUAACUGGCA------------AAUGGCGUGGUAUCUUCGCCCGCUAGAU----------------- (((((.((((((......))))))..)))))...((((......)))).------------..(((((.(((......))))))))...----------------- ( -26.20, z-score = -3.78, R) >droYak2.chr2R 19443681 106 + 21139217 UGACUUGACUGACACUCCUUAGUCUUAGUCAUUAGCCAACUGGCAAACGGGCAAACUGGCAAAUGUCUUGGUAUCUUCGCCCGCCAUUUGUGUGGGCUAGAUAAAU .(((.(((((((.(((....))).)))))))...((((....((......))....))))....)))....(((((..(((((((....).)))))).)))))... ( -33.50, z-score = -2.37, R) >droEre2.scaffold_4845 20829327 90 + 22589142 UGACUUGAGUGACACUCCUUAGUCUUAGUCAUUAGCCAACUGACAAACG----------------UCUUGGUAUCUUCGCCCGCCAUUUGUGUGGGCUAGAUAGAU .((((.(((.....)))...))))...(((....(((((..(((....)----------------)))))))((((..(((((((....).)))))).)))).))) ( -26.50, z-score = -2.02, R) >consensus UGACUUGACUGACACUCCUUAGUCUUAGUCAUUAGCCAACUAACUGGCA____________AAUGGCUUGGUAUCUUCGCCCGCCAUUUGUGUGGGCUAGAU____ (((((.((((((......))))))..)))))...((((......))))...............((((..(((......))).)))).................... (-12.28 = -14.24 + 1.96)

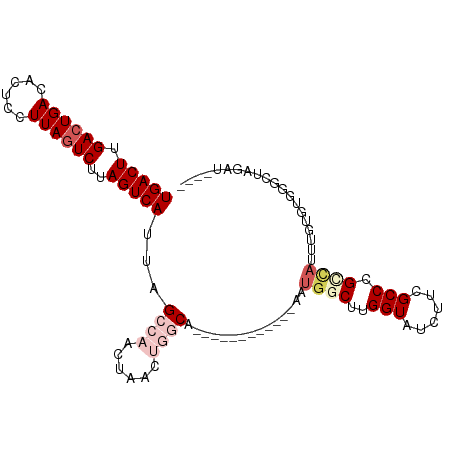

| Location | 19,469,287 – 19,469,377 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.18 |
| Shannon entropy | 0.38049 |
| G+C content | 0.46169 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -16.69 |
| Energy contribution | -15.37 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.709612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19469287 90 - 21146708 ----AUCUAGCCCACACAAAUGACGGGCGAAGAUACCAAGCCAUU------------UGCCAGUUAGUUGGCUAAUGACUAAGACUAAGGAGUGUCAGUCAAGUCA ----...(((((.((.....((((.((((((............))------------)))).)))))).))))).(((((.(.(((....))).).)))))..... ( -23.30, z-score = -1.28, R) >droSim1.chr2R 18040539 77 - 19596830 -----------------AUCUAGCGGGCGAAGAUACCACGCCAUU------------UGCCAGUUAGUUGGCUAAUGACUAAGACUAAGGAGUGUCAGUCAAGUCA -----------------..(((((.((((((............))------------)))).))))).(((((..(((((.(.(((....))).).)))))))))) ( -22.40, z-score = -1.62, R) >droSec1.super_9 2750511 77 - 3197100 -----------------AUCUAGCGGGCGAAGAUACCACGCCAUU------------UGCCAGUUAGUUGGCUAAUGACUAAGACUAAGGAGUGUCAGUCAAGUCA -----------------..(((((.((((((............))------------)))).))))).(((((..(((((.(.(((....))).).)))))))))) ( -22.40, z-score = -1.62, R) >droYak2.chr2R 19443681 106 - 21139217 AUUUAUCUAGCCCACACAAAUGGCGGGCGAAGAUACCAAGACAUUUGCCAGUUUGCCCGUUUGCCAGUUGGCUAAUGACUAAGACUAAGGAGUGUCAGUCAAGUCA ...(((((.((((.(.(....)).))))..)))))....(((....((((..(.((......)).)..))))...(((((.(.(((....))).).))))).))). ( -30.40, z-score = -1.56, R) >droEre2.scaffold_4845 20829327 90 - 22589142 AUCUAUCUAGCCCACACAAAUGGCGGGCGAAGAUACCAAGA----------------CGUUUGUCAGUUGGCUAAUGACUAAGACUAAGGAGUGUCACUCAAGUCA ...(((((.((((.(.(....)).))))..)))))...((.----------------((((.(((....))).)))).))..((((...(((.....))).)))). ( -23.00, z-score = -0.85, R) >consensus ____AUCUAGCCCACACAAAUGGCGGGCGAAGAUACCAAGCCAUU____________UGCCAGUUAGUUGGCUAAUGACUAAGACUAAGGAGUGUCAGUCAAGUCA ....................((((.((((............................)))).))))..(((((..(((((.(.(((....))).).)))))))))) (-16.69 = -15.37 + -1.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:46:01 2011