| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,464,107 – 19,464,201 |
| Length | 94 |
| Max. P | 0.980693 |

| Location | 19,464,107 – 19,464,201 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 62.82 |
| Shannon entropy | 0.71390 |
| G+C content | 0.41141 |
| Mean single sequence MFE | -18.88 |
| Consensus MFE | -9.62 |
| Energy contribution | -9.53 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
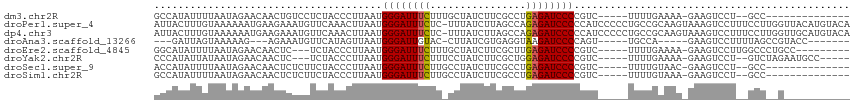
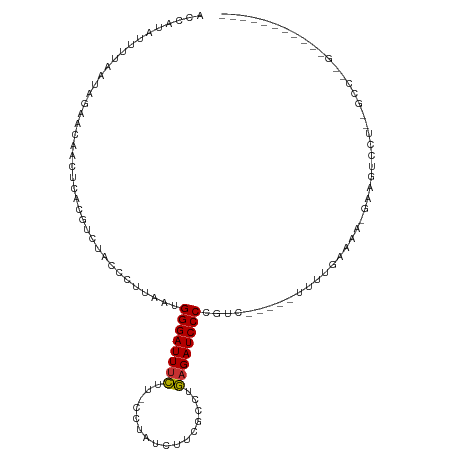
>dm3.chr2R 19464107 94 + 21146708 GCCAUAUUUUAAUAGAACAACUGUCCUCUACCCUUAAUGGGAUUUCUUUGCUAUCUUCGCCUGAGAUCCCCGUC-----UUUUGAAAA-GAAGUCCU--GCC-------------- ............((((((....))..))))........((((((((...((.......))..)))))))).(.(-----((((....)-)))).)..--...-------------- ( -17.20, z-score = -1.37, R) >droPer1.super_4 2296081 115 - 7162766 AUUACUUUGUAAAAAAUGAAGAAAUGUUCAAACUUAAUGGGAUUUCUC-UUUAUCUUAGCCAGAGAUCCCCCAUCCCCCUGCCGCAAGUAAAGUCCUUUCCUUGGUUACAUGUACA ..(((..(((((..((.((((.................((((((((((-(.......))..)))))))))........((..(....)...))...)))).))..))))).))).. ( -18.90, z-score = -0.43, R) >dp4.chr3 10236378 115 - 19779522 AUUACUUUGUAAAAAAUGAAGAAAUGUUCAAACUUAAUGGGAUUUCUC-UUUAUCUUAGCCAGAGAUCCCCCAUCCCCCUGCCGCAAGUAAAGUCCUUUCCUUGGUUGCAUGUACA ....(((..(.....)..)))...(((.((........((((((((((-(.......))..))))))))).............((((.(((.(......).))).)))).)).))) ( -19.80, z-score = -0.49, R) >droAna3.scaffold_13266 8091357 92 + 19884421 ---GAUUAGUAAAAAG---AGAAAUGUUCAUAGUUAAUGGGAUUGUAC-CUUAUCGUGAGGUAAGAUCCCCAGU-----UGCCA-----GAAGUCCUUUUAGCCGUACC------- ---.....((.(((((---.((....(((...((....((((((.(((-(((.....)))))).))))))....-----.))..-----))).))))))).))......------- ( -20.30, z-score = -2.15, R) >droEre2.scaffold_4845 20824156 98 + 22589142 GGCAUAUUUUAAUAGAACAACUC---UCUACCCUUAAUGGGAUUUCUUUGCUAUCUUCGCUUGAGAUCCCCGUC-----UUUUGAAAA-GAAGUCCUUGGCCCUGCC--------- ((((........((((.......---)))).((.....((((((((...((.......))..)))))))).(.(-----((((....)-)))).)...))...))))--------- ( -22.90, z-score = -1.80, R) >droYak2.chr2R 19438416 100 + 21139217 CCCAUAUUAUAAUAGAACAACUC---UCUACCCUUAAUGGGAUUUCUUUCCUAUCUUCGCUGGAGAUCCCCGUC-----UUUUGAAAA-GAAGUCCU--GUCUAGAAUGCC----- .......(((..((((.((..((---((((.(....((((((......))))))....).)))))).....(.(-----((((....)-)))).).)--)))))..)))..----- ( -18.60, z-score = -0.70, R) >droSec1.super_9 2745341 94 + 3197100 ACCAUAUUUUAAUAGAACAACUCUCUUCUACCCUUAAUGGGAUUUCUUGCCUAUCUUCGCCUGAGAUCCCCGUC-----UUUUGUAAC-GAAGUCCU--GCC-------------- ............(((((........)))))........((((((((..((........))..)))))))).(.(-----(((......-)))).)..--...-------------- ( -15.80, z-score = -1.57, R) >droSim1.chr2R 18034987 94 + 19596830 GCCAUAUUUUAAUAGAACAACUCUCUUCUACCCUUAAUGGGAUUUCUUGCCUAUCUUCGCCUGAGAUCCCCGUC-----UUUUGUAAA-GAAGUCCU--GCC-------------- ............(((((........)))))........((((((((..((........))..)))))))).(.(-----((((....)-)))).)..--...-------------- ( -17.50, z-score = -1.75, R) >consensus ACCAUAUUUUAAUAGAACAACUCACGUCUACCCUUAAUGGGAUUUCUU_CCUAUCUUCGCCUGAGAUCCCCGUC_____UUUUGAAAA_GAAGUCCU__GCC__G___________ ......................................((((((((................)))))))).............................................. ( -9.62 = -9.53 + -0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:59 2011