| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,449,015 – 19,449,125 |
| Length | 110 |
| Max. P | 0.774253 |

| Location | 19,449,015 – 19,449,125 |
|---|---|
| Length | 110 |
| Sequences | 13 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 70.91 |
| Shannon entropy | 0.65069 |
| G+C content | 0.52347 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -13.96 |
| Energy contribution | -13.52 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
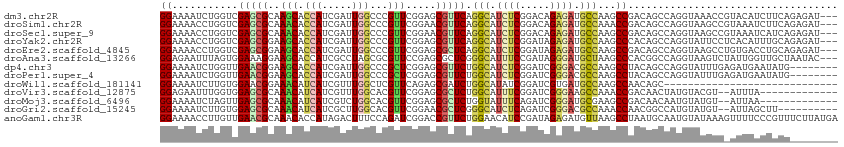
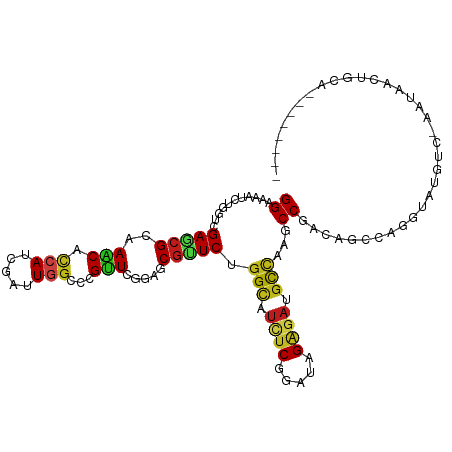
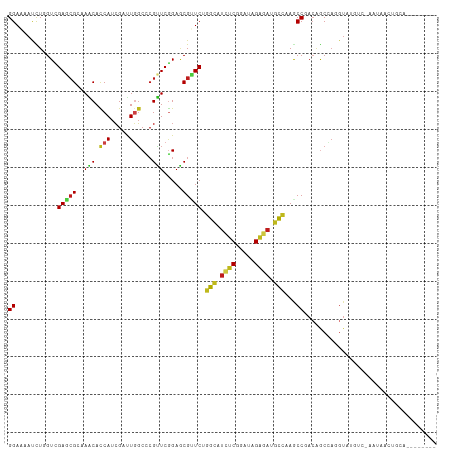
>dm3.chr2R 19449015 110 + 21146708 GGAAAAUCUGGUCGAGCGCAAGCACCAUCGAUUGGCCCGUUCGGAGCGUUCAGGCAUCUCGGACAGAGAUGCCAAGCCGACAGCCAGGUAAACCGUACAUCUUCAGAGAU--- ((...(((((((.((((((.....(((.....))).((....)).)))))).((((((((.....)))))))).........)))))))...))....((((....))))--- ( -39.60, z-score = -2.56, R) >droSim1.chr2R 18019052 110 + 19596830 GGAAAACCUGGUCGAGCGCAAACACCAUCGAUUGGCCCGUUCGGAACGUUCAGGCAUCUCGGACAGAGAUGCCAAACCGACAGCCAGGUAAGCCGUAAAUCUUCAGAGAU--- ((...(((((((.(((((......(((.....))).((....))..))))).((((((((.....)))))))).........)))))))...))....((((....))))--- ( -38.90, z-score = -3.07, R) >droSec1.super_9 2730039 110 + 3197100 GGAAAACCUGGUCGAGCGCAAACACCAUCGAUUGGCCCGUUCGGAACGUUCAGGCAUCUCGGACAGAGAUGCCAAGCCGACAGCCAGGUAAGCCGUAAAUCAUCAGAGAU--- ((...(((((((.(((((......(((.....))).((....))..))))).((((((((.....)))))))).........)))))))...))................--- ( -38.60, z-score = -2.97, R) >droYak2.chr2R 19422830 110 + 21139217 GGAAAACCUGGUCGAGCGGAAGCACCAUCGAUUGGCCCGUUCGGAGCGUUCAGGCAUCUCGGAUAGAGAUGCCAAGCCCACAGCCAGGUAUUCCUCACAUUUGCAGAGAU--- ((((.(((((((.((((((.....(((.....))).))))))((.((.....((((((((.....))))))))..))))...))))))).))))................--- ( -46.00, z-score = -4.13, R) >droEre2.scaffold_4845 20808829 110 + 22589142 GGAAAACCUGGUCGAGCGGAAGCACCAUCGAUUGGCCCGUUCGGAGCGCUCAGGCAUCUCGGAUAGAGAUGCCAAGCCGACAGCCAGGUAAGCCUGUGACCUGCAGAGAU--- .....(((((((.((((((.....(((.....))).))))))(..(.(((..((((((((.....)))))))).))))..).)))))))....((((.....))))....--- ( -43.80, z-score = -2.32, R) >droAna3.scaffold_13266 8075926 110 + 19884421 GGAGAAUUUAGUGGAAAGGAAGCACCAUCGCCUAGCGCGUUCCGAGCGCUCGGGCAUUUCCGAUAGGGAUGCUAAGCCCACGGCCAGGUAAGUCUAUUGGUUGCUAAUAC--- .......((((..(...((.....))...(((((((((.......))))).))))....((((((((..((((..(((...)))..))))..)))))))))..))))...--- ( -35.60, z-score = -0.51, R) >dp4.chr3 10220449 105 - 19779522 GGAAAAUCUGGUUGAACGGAAGCACCAUCGAUUGGCCCGCUCGGAGCGUUCUGGCAUCUCGGAUCGGGACGCCAAGCCUACAGCCAGGUAUUUGAGAUGAAUAUG-------- .(((.(((((((((...((.....)).......(((.(((.....)))...((((.(((((...))))).)))).)))..))))))))).)))............-------- ( -32.40, z-score = -0.20, R) >droPer1.super_4 2280127 105 - 7162766 GGAAAAUCUGGUUGAACGGAAGCACCAUCGAUUGGCCCGCUCGGAGCGUUCUGGCAUCUCGGAUCGGGACGCCAAGCCUACAGCCAGGUAUUUGAGAUGAAUAUG-------- .(((.(((((((((...((.....)).......(((.(((.....)))...((((.(((((...))))).)))).)))..))))))))).)))............-------- ( -32.40, z-score = -0.20, R) >droWil1.scaffold_181141 4215616 84 + 5303230 GGAAAAUCUUGUGGAACGGAAACAUCAUCGUUUGGCUCGUUCAGAGCGAUCUGGCAUAUCGGAUCGUGAUGCCAAGCCAACAGC----------------------------- ........((((((...((...((((((......((((.....))))(((((((....)))))))))))))))...)).)))).----------------------------- ( -23.80, z-score = -0.74, R) >droVir3.scaffold_12875 18354488 98 + 20611582 GGAGAAUUUGGUGGAGCGCAAACAUCAUCGUUUGGCACGUUCGGAGCGCUCUGGCAUUUCGGAUCGGGAAGCCAAACCGACAACUAUGUACGU--AUUUA------------- .......((((((((((((((((......))))....(....)..)))))))(((.(..((...))..).)))..))))).............--.....------------- ( -24.10, z-score = 0.55, R) >droMoj3.scaffold_6496 12002719 98 + 26866924 GGAAAAUCUAGUUGAGCGCAAACAUCAUCGUCUGGCACGUUCGGAGCGCUCUGGUAUUUCAGAUCGGGAUGCGAAGCCGACAACAAUGUAUGU--AUUAA------------- ...........((((..(((.((((....(((.(((.....((((....))))((((..(.....)..))))...))))))....)))).)))--.))))------------- ( -21.90, z-score = 0.84, R) >droGri2.scaffold_15245 4468511 101 + 18325388 GGAAAAUCUUGUGGAGCGCAAACAUCAUCGCUUGGCACGUUCGGAACGCUCGGGCAUCUCAGAUCGGGACGCCAAACCAACGGCCAUGUAUGU--AUUAGCUU---------- .............((((....((((....((.((((.((((.((..(....)(((.((((.....)))).)))...)))))))))).))))))--....))))---------- ( -26.80, z-score = 0.18, R) >anoGam1.chr3R 659326 113 - 53272125 GGAAAACCUUGUUGAACGCAAACACCAUAGACUUUCCAGAUCGGACCGUUCUGGAACAUCCGAUAGAGAUGUUAAGCCUAAUGCAAUGUAUAAAGUUUUCCCGUUUCUUAUGA (((((((.((((.....(((..............((((((.((...)).))))))((((((....).))))).........))).....)))).)))))))............ ( -24.90, z-score = -1.24, R) >consensus GGAAAAUCUGGUCGAGCGCAAACACCAUCGAUUGGCCCGUUCGGAGCGUUCUGGCAUCUCGGAUAGAGAUGCCAAGCCGACAGCCAGGUAUGUC_AAUAACUGCA________ ((...........(((((..(((.(((.....)))...))).....))))).(((.((((.....)))).)))...))................................... (-13.96 = -13.52 + -0.43)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:58 2011