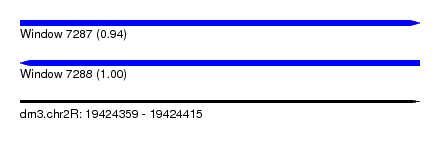
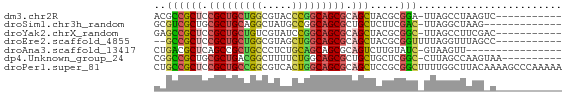
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,424,359 – 19,424,415 |
| Length | 56 |
| Max. P | 0.995370 |

| Location | 19,424,359 – 19,424,415 |
|---|---|
| Length | 56 |
| Sequences | 7 |
| Columns | 68 |
| Reading direction | forward |
| Mean pairwise identity | 61.94 |
| Shannon entropy | 0.69289 |
| G+C content | 0.64708 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -12.96 |
| Energy contribution | -13.27 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.942934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
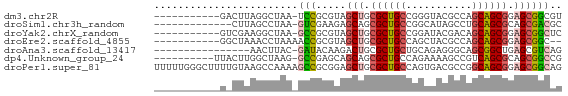
>dm3.chr2R 19424359 56 + 21146708 -----------GACUUAGGCUAA-UCCGCGUAGCUGCGCUGCCGGGUACGCCAGCAGCGGAGCGGCGU -----------......((....-.))((((.(((.((((((.((.....)).)))))).))).)))) ( -24.80, z-score = -0.54, R) >droSim1.chr3h_random 107942 54 - 1452968 -------------CUUAGCCUAA-GUCGAAGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGC -------------..........-((((.......(((((((.(((...))).)))))))..)))).. ( -22.50, z-score = -1.28, R) >droYak2.chrX_random 539170 56 + 1802292 -----------GUCGAAGGCUAA-GCCGCGUAGCUGCGCUGCCGGAUACGACAGCAGCGGAGCGGCUC -----------...........(-(((((....((((((((.((....)).)))).)))).)))))). ( -25.30, z-score = -1.23, R) >droEre2.scaffold_4855 7559 55 + 242605 -----------GGCUAAACCUAAAACCGCGUAGCUGCGCUGCCAGCUACGCCAGCAGCGGAGCGGC-- -----------..............(((((((((((......)))))))((.....))...)))).-- ( -18.60, z-score = 0.55, R) >droAna3.scaffold_13417 2534365 51 - 6960332 ----------------AACUUAC-GAUACAAGACUGCGCUGCUGCAGAGGGCAGCGGCUGAGCGUCAG ----------------.......-.......(((.((((((((((.....))))))))...))))).. ( -19.00, z-score = -1.55, R) >dp4.Unknown_group_24 89670 57 - 109830 ----------UUACUUGGCUAAG-GCCGAGCAGCAGCGCUGCCAGAAAAGCCGUCAGCGCAGCGGCCG ----------...((((((....-))))))..((.((((((.(.(.....).).)))))).))..... ( -21.10, z-score = -0.04, R) >droPer1.super_81 267490 68 - 277874 UUUUUGGGCUUUUGUAAGCCAAAAGCCGCGGAGCUGCGCUGCCAGUGACGCCGGCAGCGGAGCGGCAG ......((((((((.....))))))))((...(((.(((((((.(.....).))))))).))).)).. ( -33.90, z-score = -2.20, R) >consensus ___________G_CUAAGCCUAA_GCCGCGUAGCUGCGCUGCCGGAUACGCCAGCAGCGGAGCGGCAG ........................(((.....(((.((((((...........)))))).)))))).. (-12.96 = -13.27 + 0.31)
| Location | 19,424,359 – 19,424,415 |
|---|---|
| Length | 56 |
| Sequences | 7 |
| Columns | 68 |
| Reading direction | reverse |
| Mean pairwise identity | 61.94 |
| Shannon entropy | 0.69289 |
| G+C content | 0.64708 |
| Mean single sequence MFE | -24.04 |
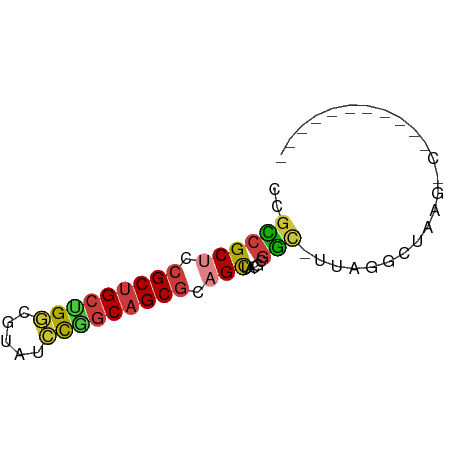
| Consensus MFE | -17.38 |
| Energy contribution | -17.29 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.995370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
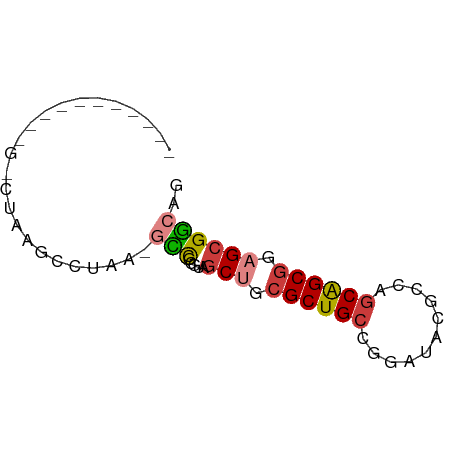
>dm3.chr2R 19424359 56 - 21146708 ACGCCGCUCCGCUGCUGGCGUACCCGGCAGCGCAGCUACGCGGA-UUAGCCUAAGUC----------- .(((.(((.(((((((((.....))))))))).)))...)))..-............----------- ( -24.60, z-score = -1.39, R) >droSim1.chr3h_random 107942 54 + 1452968 GCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCUUCGAC-UUAGGCUAAG------------- ..(((((.(((((((.(((...))).))))))).)).....)))-..........------------- ( -22.50, z-score = -0.50, R) >droYak2.chrX_random 539170 56 - 1802292 GAGCCGCUCCGCUGCUGUCGUAUCCGGCAGCGCAGCUACGCGGC-UUAGCCUUCGAC----------- (((((((...((((((((((....)))))..)))))...)))))-))..........----------- ( -25.70, z-score = -2.09, R) >droEre2.scaffold_4855 7559 55 - 242605 --GCCGCUCCGCUGCUGGCGUAGCUGGCAGCGCAGCUACGCGGUUUUAGGUUUAGCC----------- --((.(((.((((((..((...))..)))))).)))...))((((........))))----------- ( -24.00, z-score = -0.55, R) >droAna3.scaffold_13417 2534365 51 + 6960332 CUGACGCUCAGCCGCUGCCCUCUGCAGCAGCGCAGUCUUGUAUC-GUAAGUU---------------- (((.((((.....(((((.....)))))))))))).........-.......---------------- ( -14.00, z-score = -0.15, R) >dp4.Unknown_group_24 89670 57 + 109830 CGGCCGCUGCGCUGACGGCUUUUCUGGCAGCGCUGCUGCUCGGC-CUUAGCCAAGUAA---------- .(((.((.((((((.(((.....))).)))))).)).))).(((-....)))......---------- ( -23.50, z-score = -0.31, R) >droPer1.super_81 267490 68 + 277874 CUGCCGCUCCGCUGCCGGCGUCACUGGCAGCGCAGCUCCGCGGCUUUUGGCUUACAAAAGCCCAAAAA ..((.(((.(((((((((.....))))))))).)))...))((((((((.....))))))))...... ( -34.00, z-score = -3.60, R) >consensus CCGCCGCUCCGCUGCUGGCGUAUCCGGCAGCGCAGCUACGCGGC_UUAGGCUAAG_C___________ ..((((((.(((((((((.....))))))))).))).....)))........................ (-17.38 = -17.29 + -0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:54 2011