
| Sequence ID | dm3.chr2R |
|---|---|
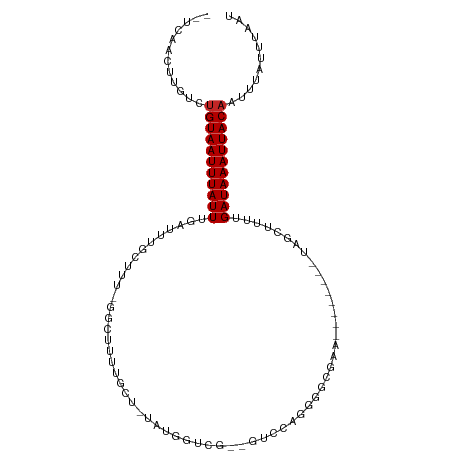
| Location | 19,388,851 – 19,388,944 |
| Length | 93 |
| Max. P | 0.838028 |

| Location | 19,388,851 – 19,388,944 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 69.47 |
| Shannon entropy | 0.62818 |
| G+C content | 0.33352 |
| Mean single sequence MFE | -21.22 |
| Consensus MFE | -5.46 |
| Energy contribution | -5.56 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.838028 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 19388851 93 + 21146708 --UCAACUUGUCUGUAAUUUAUUUGAUUUGCUCU-GGCUUUCGCU-UGUGGUCG--GUCCAGGGGCGAA--------UAGCUUUUGAUAAAUUACAAUUUAUUUAAU --..........(((((((((((...........-((((((((((-(.(((...--..))).)))))))--------.))))...)))))))))))........... ( -25.74, z-score = -3.49, R) >droSim1.chr2R 17977486 93 + 19596830 --UCAACUUGUCUGUAAUUUAUUUGAUUUGCUCU-GGCUUUCGCU-UGUGGUCG--GUCCAGGGGCGAA--------UAGCUUUUGAUAAAUUACAAUUUAUUUAAU --..........(((((((((((...........-((((((((((-(.(((...--..))).)))))))--------.))))...)))))))))))........... ( -25.74, z-score = -3.49, R) >droSec1.super_9 2677746 93 + 3197100 --UCAACUUGUCUGUAAUUUAUUUGAUUUGCUCU-GGCUUUCGCU-UGUGGUCG--GUCCAGGGGCGAA--------UAGCUUUUGAUAAAUUACAAUUUAUUUAAU --..........(((((((((((...........-((((((((((-(.(((...--..))).)))))))--------.))))...)))))))))))........... ( -25.74, z-score = -3.49, R) >droYak2.chr2R 19356646 96 + 21139217 --UCAACUUGUCUGUAAUUUAUUUGAUUUGCUUU-GGCUUCUGCUUGUUGGUUGUGGUCCAGGGGCGAA--------UAGCUUUUGAUAAAUUACAAUUUAUUUAAU --..........(((((((((((.((...(((((-.(((((((.(..(.....)..)..))))))).))--------.))).)).)))))))))))........... ( -22.30, z-score = -1.99, R) >droEre2.scaffold_4845 20754683 97 + 22589142 --UCAACUUGUCCGUAAUUUAUUUGAUUUGCUUU-GGCCGCUGCCUGUUGGCUGUGGUCCAGGGGCGAAA-------GAGCUUUUGAUAAAUUACAAUUUAUUUAAU --...........((((((((((.((....((((-((((((.(((....))).)))).))))))((....-------..)).)).))))))))))............ ( -29.30, z-score = -2.67, R) >droAna3.scaffold_13266 8027511 103 + 19884421 --UCAACUUGUCUGUAAUUUAUUUGAUUUGCUUUUGGCUUGUGUUGUAUGGUUG--GGGGAGUCGCGUUGUCCCGCUUGGCUUUUGAUAAAUUACAAUUUAUUUAAU --..........(((((((((((.((....((((..(((..........)))..--))))(((((((......)))..)))))).)))))))))))........... ( -19.70, z-score = -0.19, R) >dp4.chr3 10771633 71 - 19779522 --UCAACUUCUCUGUAAUUUAUUUGAUUUGCUUU------UUGCCGCAAGGCCG----------------------------UUUGAUAAAUUACAAUUUAUUUAAU --..........(((((((((((.(((.......------..(((....))).)----------------------------)).)))))))))))........... ( -14.60, z-score = -2.85, R) >droPer1.super_4 2833054 71 - 7162766 --UCAACUUCUCUGUAAUUUAUUUGAUUUGCUUU------UUGCCGCAAGGCCG----------------------------UUUGAUAAAUUACAAUUUAUUUAAU --..........(((((((((((.(((.......------..(((....))).)----------------------------)).)))))))))))........... ( -14.60, z-score = -2.85, R) >droMoj3.scaffold_6496 12351331 75 + 26866924 GCUCGAGUGCUGUGUAAUUUAUUUGAUUUGCUUUCGCAUC--------------------------GCUGU------UGCUCAAUGAUAAAUUACAAUUUAUUUAAU ....(((((...((((((((((((((..(((....)))..--------------------------((...------.)))))..)))))))))))...)))))... ( -14.80, z-score = -1.10, R) >droGri2.scaffold_15245 4781786 99 + 18325388 --UCAAGUGCUAUGUAAUUUAUUUGACUUGCUUUCGUUUCAAUACGUUUCAUUUAGCCUUUGGCUUGCGAC------UGCACAUUGAUAAAUUACAAUUUAUUUAAU --..(((((...(((((((((((.....(((..((((.((((...(((......)))..))))...)))).------.)))....)))))))))))...)))))... ( -19.70, z-score = -2.04, R) >consensus __UCAACUUGUCUGUAAUUUAUUUGAUUUGCUUU_GGCUUUUGCU_UAUGGUCG__GUCCAGGGGCGAA________UAGCUUUUGAUAAAUUACAAUUUAUUUAAU ............(((((((((((..............................................................)))))))))))........... ( -5.46 = -5.56 + 0.10)


| Location | 19,388,851 – 19,388,944 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 69.47 |
| Shannon entropy | 0.62818 |
| G+C content | 0.33352 |
| Mean single sequence MFE | -15.54 |
| Consensus MFE | -4.17 |
| Energy contribution | -4.28 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.656687 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 19388851 93 - 21146708 AUUAAAUAAAUUGUAAUUUAUCAAAAGCUA--------UUCGCCCCUGGAC--CGACCACA-AGCGAAAGCC-AGAGCAAAUCAAAUAAAUUACAGACAAGUUGA-- ..........(((((((((((.....(((.--------(((((...(((..--...)))..-.)))))))).-.((.....))..))))))))))).........-- ( -19.00, z-score = -3.76, R) >droSim1.chr2R 17977486 93 - 19596830 AUUAAAUAAAUUGUAAUUUAUCAAAAGCUA--------UUCGCCCCUGGAC--CGACCACA-AGCGAAAGCC-AGAGCAAAUCAAAUAAAUUACAGACAAGUUGA-- ..........(((((((((((.....(((.--------(((((...(((..--...)))..-.)))))))).-.((.....))..))))))))))).........-- ( -19.00, z-score = -3.76, R) >droSec1.super_9 2677746 93 - 3197100 AUUAAAUAAAUUGUAAUUUAUCAAAAGCUA--------UUCGCCCCUGGAC--CGACCACA-AGCGAAAGCC-AGAGCAAAUCAAAUAAAUUACAGACAAGUUGA-- ..........(((((((((((.....(((.--------(((((...(((..--...)))..-.)))))))).-.((.....))..))))))))))).........-- ( -19.00, z-score = -3.76, R) >droYak2.chr2R 19356646 96 - 21139217 AUUAAAUAAAUUGUAAUUUAUCAAAAGCUA--------UUCGCCCCUGGACCACAACCAACAAGCAGAAGCC-AAAGCAAAUCAAAUAAAUUACAGACAAGUUGA-- ..........(((((((((((.....(((.--------(((((...(((.......)))....)).)))...-..))).......))))))))))).........-- ( -12.30, z-score = -1.38, R) >droEre2.scaffold_4845 20754683 97 - 22589142 AUUAAAUAAAUUGUAAUUUAUCAAAAGCUC-------UUUCGCCCCUGGACCACAGCCAACAGGCAGCGGCC-AAAGCAAAUCAAAUAAAUUACGGACAAGUUGA-- ..........(((((((((((.....((..-------....))...(((.((.(.(((....))).).))))-)...........))))))))))).........-- ( -18.50, z-score = -0.92, R) >droAna3.scaffold_13266 8027511 103 - 19884421 AUUAAAUAAAUUGUAAUUUAUCAAAAGCCAAGCGGGACAACGCGACUCCCC--CAACCAUACAACACAAGCCAAAAGCAAAUCAAAUAAAUUACAGACAAGUUGA-- ..........(((((((((((..........(((......)))........--................((.....)).......))))))))))).........-- ( -13.70, z-score = -1.73, R) >dp4.chr3 10771633 71 + 19779522 AUUAAAUAAAUUGUAAUUUAUCAAA----------------------------CGGCCUUGCGGCAA------AAAGCAAAUCAAAUAAAUUACAGAGAAGUUGA-- ..........(((((((((((....----------------------------..(..((((.....------...))))..)..))))))))))).........-- ( -10.30, z-score = -0.86, R) >droPer1.super_4 2833054 71 + 7162766 AUUAAAUAAAUUGUAAUUUAUCAAA----------------------------CGGCCUUGCGGCAA------AAAGCAAAUCAAAUAAAUUACAGAGAAGUUGA-- ..........(((((((((((....----------------------------..(..((((.....------...))))..)..))))))))))).........-- ( -10.30, z-score = -0.86, R) >droMoj3.scaffold_6496 12351331 75 - 26866924 AUUAAAUAAAUUGUAAUUUAUCAUUGAGCA------ACAGC--------------------------GAUGCGAAAGCAAAUCAAAUAAAUUACACAGCACUCGAGC ...........((((((((((..((((((.------...))--------------------------..(((....)))..))))))))))))))............ ( -16.50, z-score = -3.08, R) >droGri2.scaffold_15245 4781786 99 - 18325388 AUUAAAUAAAUUGUAAUUUAUCAAUGUGCA------GUCGCAAGCCAAAGGCUAAAUGAAACGUAUUGAAACGAAAGCAAGUCAAAUAAAUUACAUAGCACUUGA-- ...........(((((((((((((((((..------.(((..((((...))))...)))..))))))))..(....).........)))))))))..........-- ( -16.80, z-score = -0.87, R) >consensus AUUAAAUAAAUUGUAAUUUAUCAAAAGCUA________UUCGCCCCUGGAC__CGACCAAA_AGCAAAAGCC_AAAGCAAAUCAAAUAAAUUACAGACAAGUUGA__ ..........(((((((((((................................................................)))))))))))........... ( -4.17 = -4.28 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:51 2011