| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,384,300 – 19,384,538 |
| Length | 238 |
| Max. P | 0.869000 |

| Location | 19,384,300 – 19,384,425 |
|---|---|
| Length | 125 |
| Sequences | 6 |
| Columns | 140 |
| Reading direction | forward |
| Mean pairwise identity | 70.04 |
| Shannon entropy | 0.56259 |
| G+C content | 0.50410 |
| Mean single sequence MFE | -41.03 |
| Consensus MFE | -16.52 |
| Energy contribution | -18.58 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.628705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
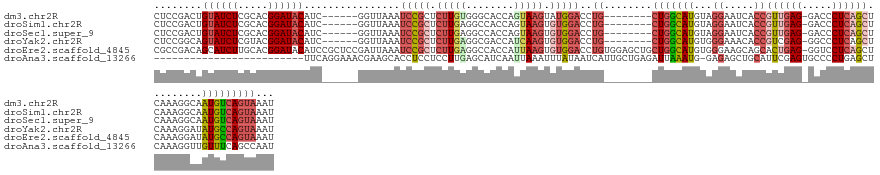
>dm3.chr2R 19384300 125 + 21146708 CUCCGACUGUAUCUCGCACGGAUACAUC------GGUUAAAUCCGCUCUUGUGGGCACCAGUAAGUAUGGACCUG--------CUGGCAUGUAGGAAUCACCGUUGAG-GACCCUCAGCUCAAAGGCAAUGUCAGUAAAU ..((((.(((((((.....)))))))))------)).....((((..(((((((...))).))))..))))..((--------(((((((((.((.....))((((((-....))))))......)).)))))))))... ( -40.90, z-score = -1.79, R) >droSim1.chr2R 17972895 125 + 19596830 CUCCGACUGUAUCUCGCACGGAUACAUC------GGUUAAAUCCGCUCUUGAGGCCACCAGUAAGUGUGGACCUG--------CUGGCAUGUAGGAAUCACCGUUGAG-GACCCUCAGCUCAAAGGCAAUGUCAGUAAAU ..((((.(((((((.....)))))))))------)).....(((((.((((..(....)..)))).)))))..((--------(((((((((.((.....))((((((-....))))))......)).)))))))))... ( -42.60, z-score = -2.20, R) >droSec1.super_9 2673181 125 + 3197100 CUCCGACUGUAUCUCGCACGGAUACAUC------GGUUAAAUCCGCUCUUGAGGCCACCAGUAAGUGUGGACCUG--------CUGGCAUGUAGGAAUCACCGUUGAG-GACCCUCAGCUCAAAGGCAAUGUCAGUAAAU ..((((.(((((((.....)))))))))------)).....(((((.((((..(....)..)))).)))))..((--------(((((((((.((.....))((((((-....))))))......)).)))))))))... ( -42.60, z-score = -2.20, R) >droYak2.chr2R 19352276 125 + 21139217 CUCCGGCAGUAUCUCGUACGGAUACAUC------GGUUAAAUCCGCUCUUGAGGCGACCAUCAAGUGUGGACCUG--------CUGGCAUGUGGGAAACACCGUCGAG-GGCCCUCAGCUCAAAGGAUAUGCCAGUAAAU ..((((..((((((.....)))))).))------)).....(((((.(((((((...)).))))).)))))..((--------(((((((((((......))((.(((-....))).)).......)))))))))))... ( -45.30, z-score = -1.99, R) >droEre2.scaffold_4845 20750216 139 + 22589142 CGCCGACAGCAUCUUGCACGGAUACAUCCGCUCCGAUUAAAUCCGCUCUUGAGGCCACCAUUAAGUGUGGACCUGUGGAGCUGCUGGCAUGUGGGAAGCAGCACUGAG-GGUCCUCAGCUCAAAGGAUAUGCCAGUAAAU ..(((...((.....)).)))........((((((......(((((.(((((((...)).))))).)))))....))))))(((((((((((.(....)....(((((-....)))))........)))))))))))... ( -47.60, z-score = -0.68, R) >droAna3.scaffold_13266 8023142 114 + 19884421 -------------------------UUCAGGAAACGAAGCACCUCCUCCUUGAGCAUCAAUUAAAUUUAUAAUCAUUGCUGAGAUUAAAUG-GAGAGCUGCAUUCGAGUGCCCCUGAGCUCAAAGGUUGUUUCAGCCAAU -------------------------((((((...(((((((.(((.(((((.((((...((((......))))...)))).)).......)-))))).))).))))......))))))......(((((...)))))... ( -27.21, z-score = -0.59, R) >consensus CUCCGACUGUAUCUCGCACGGAUACAUC______GGUUAAAUCCGCUCUUGAGGCCACCAGUAAGUGUGGACCUG________CUGGCAUGUAGGAAUCACCGUUGAG_GACCCUCAGCUCAAAGGCAAUGUCAGUAAAU ........((((((.....))))))................(((((.(((((........))))).)))))............(((((((...((.....))((((((.....)))))).........)))))))..... (-16.52 = -18.58 + 2.06)
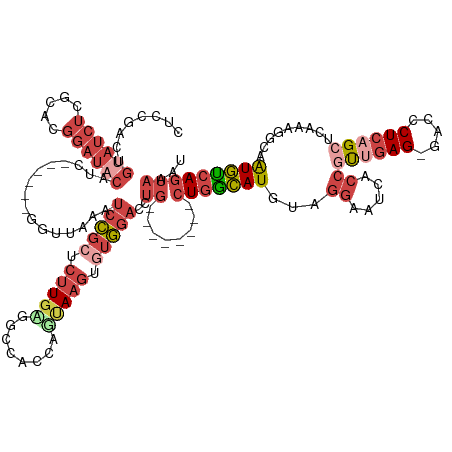
| Location | 19,384,425 – 19,384,538 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 92.99 |
| Shannon entropy | 0.12061 |
| G+C content | 0.49297 |
| Mean single sequence MFE | -38.72 |
| Consensus MFE | -31.30 |
| Energy contribution | -31.66 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.869000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19384425 113 + 21146708 GAGAGUUUAACAGCAGGGAUGAAGAGGCAAGUAGAGUGCCAAUUACUGCUUCACUUACCUUUGAGAGUCCCAGCCAGUUGGCCCCGGAUAUAAGUCCAGC---AGUGACUCACUGA ..(((((..((.((.(((((..(((((.((((..((((........))))..)))).)))))....))))).(((....)))...((((....)))).))---.)))))))..... ( -35.80, z-score = -2.02, R) >droSim1.chr2R 17973020 113 + 19596830 GAGAGUUUAACAGCAGGGAUGAAGAGGCAAGUAGAGUGCCAAUUACUGCUUCACUUACCUUUGAGAGUCCCAGCCAGUUGGCCCCGGAUUUAAGUCCUGC---AGUGACUCACUGA ..(((((..((.((((((....(((((.((((..((((........))))..)))).)))))..((((((..(((....)))...))))))...))))))---.)))))))..... ( -36.40, z-score = -1.83, R) >droSec1.super_9 2673306 113 + 3197100 GAGAGUUUAACAGCAGGGAUGAAGAGGCAAGUAGAGUGCCAAUUACUGCUUCACUUACCUUUGAGAGUCCCAGCCAGUUGGCCCCGGAUUUAAGUCCUGC---AGUGACUCACUGA ..(((((..((.((((((....(((((.((((..((((........))))..)))).)))))..((((((..(((....)))...))))))...))))))---.)))))))..... ( -36.40, z-score = -1.83, R) >droYak2.chr2R 19352401 116 + 21139217 GAGAGUUUAACUGCAGGGAUGAAGAGGAAAGUAGAGUGCCAAUUACUGCUUCACUUACCUUUGAGUGUCCCUGCCAGUUGGCUCCGGAUUUAAGUCCUCCUGGAGUGACUCGCUGA ..(((((((((((((((((((.(((((.((((..((((........))))..)))).)))))...)))))))).))))))(((((((((....))))....))))))))))..... ( -45.60, z-score = -3.67, R) >droEre2.scaffold_4845 20750355 113 + 22589142 GAGAGUUUAACUGCGGGGAAGAAGAGGAAAGUGGAGUGCCAAUUACUGUUUCACUUACCUUUGAGAGUUCCUGCCAGUUGGCCCAGGAUUUAAGUCCUGC---AGUGACUCGCUGA ..(((((..(((((((((....(((((.(((((((..(........)..))))))).)))))......(((((((....)))..))))......))))))---))))))))..... ( -39.40, z-score = -2.15, R) >consensus GAGAGUUUAACAGCAGGGAUGAAGAGGCAAGUAGAGUGCCAAUUACUGCUUCACUUACCUUUGAGAGUCCCAGCCAGUUGGCCCCGGAUUUAAGUCCUGC___AGUGACUCACUGA ..(((((.....((((((....(((((.((((.(((.((........))))))))).)))))..((((((..(((....)))...))))))...))))))......)))))..... (-31.30 = -31.66 + 0.36)

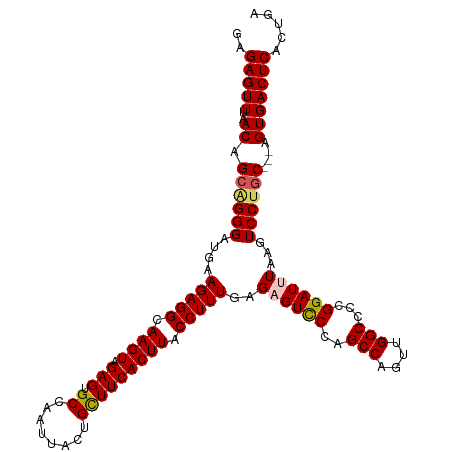
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:49 2011