
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,381,735 – 19,381,789 |
| Length | 54 |
| Max. P | 0.657910 |

| Location | 19,381,735 – 19,381,789 |
|---|---|
| Length | 54 |
| Sequences | 5 |
| Columns | 58 |
| Reading direction | forward |
| Mean pairwise identity | 67.43 |
| Shannon entropy | 0.58320 |
| G+C content | 0.50864 |
| Mean single sequence MFE | -13.44 |
| Consensus MFE | -5.94 |
| Energy contribution | -6.62 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.657910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
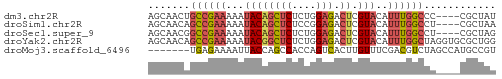
>dm3.chr2R 19381735 54 + 21146708 AGCAACUGCCGAAAAAUACAGCUCUCUGGAGACUCGUACAUUUGGCCC----CGCUAU (((....((((((...((((((((....))).)).)))..))))))..----.))).. ( -13.80, z-score = -1.84, R) >droSim1.chr2R 17963306 54 + 19596830 AGCAACAGCCGAAAAAUACAGCUCUCCGGAGACUCGUACAUUUGGCCU----CGCUAA (((....((((((...((((((((....))).)).)))..))))))..----.))).. ( -13.80, z-score = -1.72, R) >droSec1.super_9 2670664 54 + 3197100 AGCAACGGCCGAAAAAUACAGCUCUCUGGAGACUCGUACAUUUGGCCU----CGCUAG (((...(((((((...((((((((....))).)).)))..))))))).----.))).. ( -17.50, z-score = -2.68, R) >droYak2.chr2R 19349716 58 + 21139217 AGCAACAGCCGAAAAAUACGGCUCUCUGGAGACUCGUACAUUUGGCUAGGUGCGCUGG .(((..(((((((...((((..(((....)))..))))..)))))))...)))..... ( -19.40, z-score = -2.01, R) >droMoj3.scaffold_6496 12342582 51 + 26866924 -------UGAGAAAAUUACCAGCCACCAGUCACUUGUUUCGACGUCUAGCCAUGCCGU -------..............((.....(((.........))).....))........ ( -2.70, z-score = 1.70, R) >consensus AGCAACAGCCGAAAAAUACAGCUCUCUGGAGACUCGUACAUUUGGCCA____CGCUAG .......((((((...((((((((....))).)).)))..))))))............ ( -5.94 = -6.62 + 0.68)
| Location | 19,381,735 – 19,381,789 |
|---|---|
| Length | 54 |
| Sequences | 5 |
| Columns | 58 |
| Reading direction | reverse |
| Mean pairwise identity | 67.43 |
| Shannon entropy | 0.58320 |
| G+C content | 0.50864 |
| Mean single sequence MFE | -15.80 |
| Consensus MFE | -6.46 |
| Energy contribution | -7.74 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.540442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
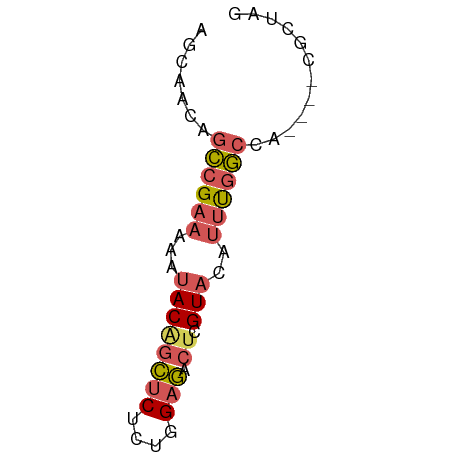
>dm3.chr2R 19381735 54 - 21146708 AUAGCG----GGGCCAAAUGUACGAGUCUCCAGAGAGCUGUAUUUUUCGGCAGUUGCU ..((((----(.((((((.((((.((.(((....))))))))).))).)))..))))) ( -15.40, z-score = -0.98, R) >droSim1.chr2R 17963306 54 - 19596830 UUAGCG----AGGCCAAAUGUACGAGUCUCCGGAGAGCUGUAUUUUUCGGCUGUUGCU ..((((----((((((((.((((.((.(((....))))))))).))).)))).))))) ( -16.70, z-score = -1.31, R) >droSec1.super_9 2670664 54 - 3197100 CUAGCG----AGGCCAAAUGUACGAGUCUCCAGAGAGCUGUAUUUUUCGGCCGUUGCU ..((((----((((((((.((((.((.(((....))))))))).))).)))).))))) ( -19.40, z-score = -2.63, R) >droYak2.chr2R 19349716 58 - 21139217 CCAGCGCACCUAGCCAAAUGUACGAGUCUCCAGAGAGCCGUAUUUUUCGGCUGUUGCU .....(((..((((((((.(((((.(.(((....))))))))).))).))))).))). ( -17.80, z-score = -2.28, R) >droMoj3.scaffold_6496 12342582 51 - 26866924 ACGGCAUGGCUAGACGUCGAAACAAGUGACUGGUGGCUGGUAAUUUUCUCA------- .((((...(((((.((.(.......))).))))).))))............------- ( -9.70, z-score = 0.51, R) >consensus AUAGCG____AGGCCAAAUGUACGAGUCUCCAGAGAGCUGUAUUUUUCGGCUGUUGCU ...........(((((((.(((((..(((....)))..))))).))).))))...... ( -6.46 = -7.74 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:46 2011