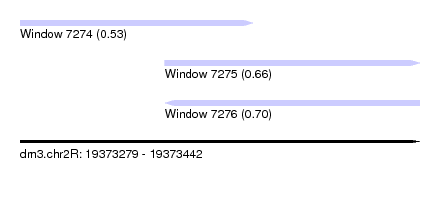
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,373,279 – 19,373,442 |
| Length | 163 |
| Max. P | 0.704349 |

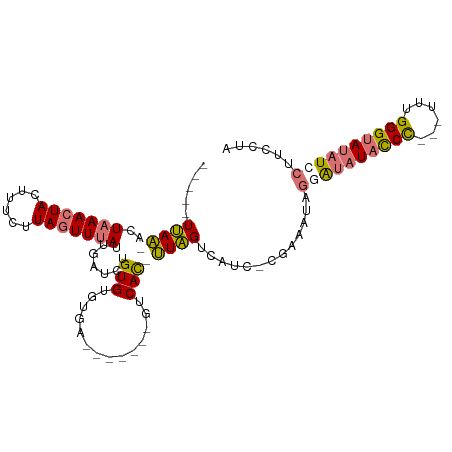
| Location | 19,373,279 – 19,373,374 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 69.08 |
| Shannon entropy | 0.46857 |
| G+C content | 0.36129 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -10.24 |
| Energy contribution | -11.55 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.525868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19373279 95 + 21146708 -----UUAA-ACUAAACUACUUUCUUAGUUUAUUGAUCUGUGUGUGA-------GUCAC-UUAGUUAUCGUGGGAUGGUAUAUACCCACAUUUGGGUAUAUCCUUCCUA -----.(((-(((((.........))))))))..((((((.(((...-------..)))-.)))..))).(((((.((.(((((((((....))))))))))).))))) ( -27.70, z-score = -3.29, R) >droSim1.chr2R 17954967 91 + 19596830 -----UUAA-GCUAAACUACUUUCUUAGUUUAUUAAUCUGUGUGUGG-------GUCAC-UUAGUCAUC-CGAACUAGGGUAUAUCC---UUUGGGUAUAUCCUUCUUA -----.(((-(((((.........)))))))).........((.(((-------((.((-...)).)))-)).)).(((((((((((---...)))))))))))..... ( -20.40, z-score = -1.58, R) >droSec1.super_9 2662045 91 + 3197100 -----UUAA-ACUAAACUACUUUCUUAGUUUAUUGAUCUGUGUGUGA-------GUCAC-UUAGUCAUC-CGAACUAGGGUAUAUCC---UUUGGGUAUAUCCGUCUUA -----....-...............((((((..(((.(((.(((...-------..)))-.))))))..-.))))))((((((((((---...))))))))))...... ( -22.70, z-score = -2.47, R) >droEre2.scaffold_4845 20739709 109 + 22589142 UCACAUCAGCAGUUAAAUACUUUUUUAAUUUAUGGGUCUGUGAGUAUCUAAGUUGUCAUGCUGGCGAUCCCCCUAUGAUAUAUACCUCCAUCUGGGCAACUCCUUACUU ((((((((..(((((((......)))))))..)))...)))))(((....(((((((....(((.(...................).)))....)))))))...))).. ( -18.31, z-score = 0.81, R) >consensus _____UUAA_ACUAAACUACUUUCUUAGUUUAUUGAUCUGUGUGUGA_______GUCAC_UUAGUCAUC_CGAAAUAGGAUAUACCC___UUUGGGUAUAUCCUUCCUA ............(((((((......))))))).....(((...((((........))))..))).............((((((((((......))))))))))...... (-10.24 = -11.55 + 1.31)


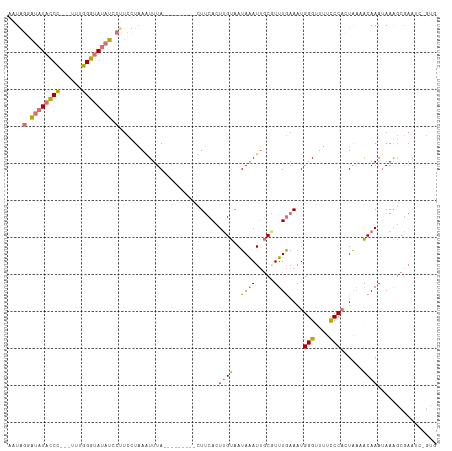
| Location | 19,373,338 – 19,373,442 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.07 |
| Shannon entropy | 0.48339 |
| G+C content | 0.34189 |
| Mean single sequence MFE | -23.25 |
| Consensus MFE | -11.50 |
| Energy contribution | -11.62 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.657914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19373338 104 + 21146708 GAUGGUAUAUACCCACAUUUGGGUAUAUCCUUCCUAAGUUUA---------UUUCAGUUGUAAUAAAUUGCAUUUGAAAUGGGGUUUUCCAUUA--ACAAA-AAAGCGAAUCUGUU ((.((.(((((((((....))))))))))).))....((((.---------..((((.(((((....))))).))))((((((....)))))).--.....-.))))......... ( -25.00, z-score = -2.16, R) >droSim1.chr2R 17955025 104 + 19596830 ACUAGGGUAUAUCC---UUUGGGUAUAUCCUUCUUAAAUUUA---------UUUCACUUGUUAUAAAUUGCGUUUGAAAUGGGUUUUCCCACUAAAACAAAUAAAGCGAAUCAGUG (((.((((((((((---...))))))))))(((...((((((---------(..(....)..)))))))..((((....((((....))))...)))).........)))..))). ( -22.80, z-score = -2.21, R) >droSec1.super_9 2662103 104 + 3197100 ACUAGGGUAUAUCC---UUUGGGUAUAUCCGUCUUAAAUUUA---------CUUCACUUGUAAUAGAUUGCGUUUGAAAUGGGUUUUCCCACUAAAACAAAUAAAGCGAAUAGGUG ....((((((((((---...)))))))))).......(((((---------.(((.(((((((....))))((((....((((....))))...)))).....))).)))))))). ( -24.90, z-score = -1.92, R) >droEre2.scaffold_4845 20739783 106 + 22589142 -AUGAUAUAUACCUCCAUCUGGGCAACUCCUUACUUAGUUUAAGGUACAUACUGCAGUAGCAAUAAAUGCCCUUUAAAAUGGGUUUUCCCUCUGAUGCAAAUAAGCC--------- -..............((((.(((((...(((((.......))))).......(((....))).....)))))........(((....)))...))))..........--------- ( -20.30, z-score = 0.05, R) >consensus AAUAGGAUAUACCC___UUUGGGUAUAUCCUUCCUAAAUUUA_________CUUCACUUGUAAUAAAUUGCGUUUGAAAUGGGUUUUCCCACUAAAACAAAUAAAGCGAAUC_GUG ....((((((((((......))))))))))...........................((((..((((.....))))....(((....)))......))))................ (-11.50 = -11.62 + 0.12)


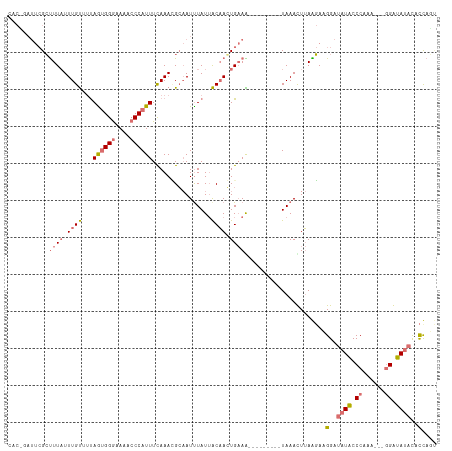
| Location | 19,373,338 – 19,373,442 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 69.07 |
| Shannon entropy | 0.48339 |
| G+C content | 0.34189 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -9.79 |
| Energy contribution | -13.48 |
| Covariance contribution | 3.69 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19373338 104 - 21146708 AACAGAUUCGCUUU-UUUGU--UAAUGGAAAACCCCAUUUCAAAUGCAAUUUAUUACAACUGAAA---------UAAACUUAGGAAGGAUAUACCCAAAUGUGGGUAUAUACCAUC ......(((((...-((((.--.(((((......))))).)))).))............((((..---------.....)))))))(((((((((((....))))))))).))... ( -24.80, z-score = -2.94, R) >droSim1.chr2R 17955025 104 - 19596830 CACUGAUUCGCUUUAUUUGUUUUAGUGGGAAAACCCAUUUCAAACGCAAUUUAUAACAAGUGAAA---------UAAAUUUAAGAAGGAUAUACCCAAA---GGAUAUACCCUAGU ......(((((((.((((((((.((((((....))))))..))))).))).......))))))).---------...........(((.((((((....---)).)))).)))... ( -18.30, z-score = -1.09, R) >droSec1.super_9 2662103 104 - 3197100 CACCUAUUCGCUUUAUUUGUUUUAGUGGGAAAACCCAUUUCAAACGCAAUCUAUUACAAGUGAAG---------UAAAUUUAAGACGGAUAUACCCAAA---GGAUAUACCCUAGU .....(((..((((((((((..(((((((....)))).............)))..))))))))))---------..))).......((.((((((....---)).)))).)).... ( -21.41, z-score = -1.88, R) >droEre2.scaffold_4845 20739783 106 - 22589142 ---------GGCUUAUUUGCAUCAGAGGGAAAACCCAUUUUAAAGGGCAUUUAUUGCUACUGCAGUAUGUACCUUAAACUAAGUAAGGAGUUGCCCAGAUGGAGGUAUAUAUCAU- ---------....(((((.((((...(((....)))........(((((..((((((....))))))....(((((.......)))))...))))).)))).)))))........- ( -30.80, z-score = -2.07, R) >consensus CAC_GAUUCGCUUUAUUUGUUUUAGUGGGAAAACCCAUUUCAAACGCAAUUUAUUACAACUGAAA_________UAAACUUAAGAAGGAUAUACCCAAA___GGAUAUACACCAGU ...........((((((((((..((((((....))))))...............))))))))))......................((((((((((......)))))))))).... ( -9.79 = -13.48 + 3.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:45 2011