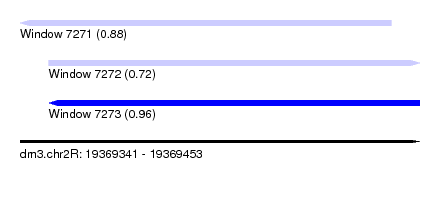
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,369,341 – 19,369,453 |
| Length | 112 |
| Max. P | 0.959346 |

| Location | 19,369,341 – 19,369,445 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 91.00 |
| Shannon entropy | 0.12362 |
| G+C content | 0.36137 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -22.31 |
| Energy contribution | -23.20 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.875934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
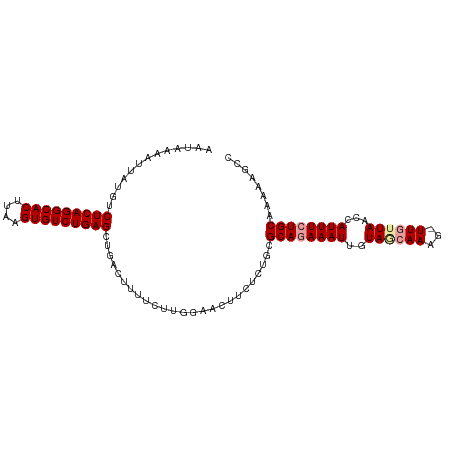
>dm3.chr2R 19369341 104 - 21146708 AAUAAAAUUAUGUCUCAGGCACUUAAGUGUCUGAGCCGAUUUUUCUUGGAAAUUGUCUGCGCACAAAUUGUAACAAAAUUUAUUAACCAUUUCUGCAAAAAGCC .............(((((((((....))))))))).(((((((.....))))))).....(((.((((.((..............)).)))).)))........ ( -19.84, z-score = -1.17, R) >droSim1.chr2R 17950737 103 - 19596830 AAUAAAAUUAUGUCUCAGGCACUUAAGUGUCUGAGUUGACUUUUCUUGGAACUUCUUUGCGCAGAAAUUGUAGCAAAG-UUGUUAACCAUUUCUGCAAAAAGCC .............(((((((((....)))))))))...................((((..((((((((..(((((...-.)))))...))))))))..)))).. ( -28.40, z-score = -2.77, R) >droSec1.super_9 2658078 103 - 3197100 AAUAAAAUUAUGUCUCAGGCACUGAAGUGUCUGAGCUGUCUUUUCUUGGAACUUCUCUGCGCAGAAAUUGUAGCAAAG-UUGAUAACCAUUUCUGCAAAAAGCC ...........(.(((((((((....))))))))))..(((......)))..........((((((((.((..((...-.))...)).))))))))........ ( -25.20, z-score = -1.34, R) >consensus AAUAAAAUUAUGUCUCAGGCACUUAAGUGUCUGAGCUGACUUUUCUUGGAACUUCUCUGCGCAGAAAUUGUAGCAAAG_UUGUUAACCAUUUCUGCAAAAAGCC .............(((((((((....))))))))).........................((((((((..(((((.....)))))...))))))))........ (-22.31 = -23.20 + 0.89)

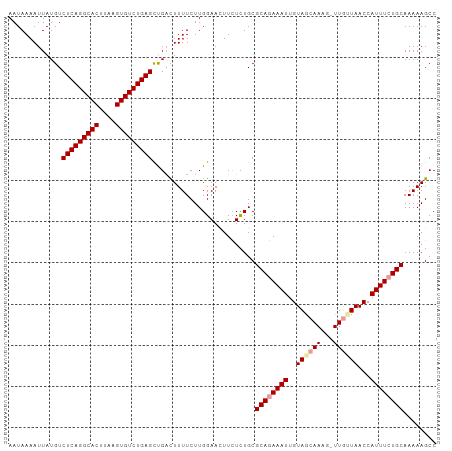
| Location | 19,369,349 – 19,369,453 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 90.35 |
| Shannon entropy | 0.13245 |
| G+C content | 0.37752 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -21.61 |
| Energy contribution | -22.17 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.721823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19369349 104 + 21146708 GCAGAAAUGGUUAAUAAAUUUUGUUACAAUUUGUGCGCAGACAAUUUCCAAGAAAAAUCGGCUCAGACACUUAAGUGCCUGAGACAUAAUUUUAUUGCGACAAA (((.((((...(((((.....)))))..)))).)))((((....((((...))))....(.(((((.(((....))).))))).).........))))...... ( -20.70, z-score = -0.92, R) >droSim1.chr2R 17950745 103 + 19596830 GCAGAAAUGGUUAACAA-CUUUGCUACAAUUUCUGCGCAAAGAAGUUCCAAGAAAAGUCAACUCAGACACUUAAGUGCCUGAGACAUAAUUUUAUUGCGGCAAA ((((((((.((...((.-...))..)).))))))))((((.((((((....((....))..(((((.(((....))).)))))....)))))).))))...... ( -26.50, z-score = -2.40, R) >droSec1.super_9 2658086 103 + 3197100 GCAGAAAUGGUUAUCAA-CUUUGCUACAAUUUCUGCGCAGAGAAGUUCCAAGAAAAGACAGCUCAGACACUUCAGUGCCUGAGACAUAAUUUUAUUGCGGCAAA ((((((((.((...((.-...))..)).))))))))........((..(((.((((.....(((((.(((....))).)))))......)))).)))..))... ( -25.60, z-score = -1.31, R) >consensus GCAGAAAUGGUUAACAA_CUUUGCUACAAUUUCUGCGCAGAGAAGUUCCAAGAAAAGUCAGCUCAGACACUUAAGUGCCUGAGACAUAAUUUUAUUGCGGCAAA ((((((((.((...((.....))..)).))))))))((((.((((((....(......)..(((((.(((....))).)))))....)))))).))))...... (-21.61 = -22.17 + 0.56)

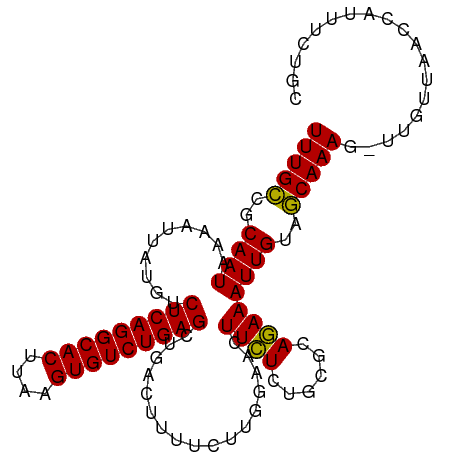
| Location | 19,369,349 – 19,369,453 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 90.35 |
| Shannon entropy | 0.13245 |
| G+C content | 0.37752 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -24.02 |
| Energy contribution | -23.13 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
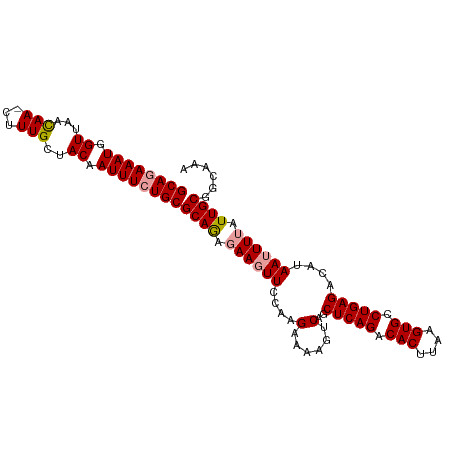
>dm3.chr2R 19369349 104 - 21146708 UUUGUCGCAAUAAAAUUAUGUCUCAGGCACUUAAGUGUCUGAGCCGAUUUUUCUUGGAAAUUGUCUGCGCACAAAUUGUAACAAAAUUUAUUAACCAUUUCUGC (((((((((((((........(((((((((....)))))))))((((......))))...)))).)))).)))))............................. ( -24.90, z-score = -2.64, R) >droSim1.chr2R 17950745 103 - 19596830 UUUGCCGCAAUAAAAUUAUGUCUCAGGCACUUAAGUGUCUGAGUUGACUUUUCUUGGAACUUCUUUGCGCAGAAAUUGUAGCAAAG-UUGUUAACCAUUUCUGC ......((((...........(((((((((....)))))))))..((..(((....)))..)).))))((((((((..(((((...-.)))))...)))))))) ( -29.40, z-score = -2.72, R) >droSec1.super_9 2658086 103 - 3197100 UUUGCCGCAAUAAAAUUAUGUCUCAGGCACUGAAGUGUCUGAGCUGUCUUUUCUUGGAACUUCUCUGCGCAGAAAUUGUAGCAAAG-UUGAUAACCAUUUCUGC ......(((..........(.(((((((((....))))))))))..(((......))).......)))((((((((.((..((...-.))...)).)))))))) ( -26.80, z-score = -1.45, R) >consensus UUUGCCGCAAUAAAAUUAUGUCUCAGGCACUUAAGUGUCUGAGCUGACUUUUCUUGGAACUUCUCUGCGCAGAAAUUGUAGCAAAG_UUGUUAACCAUUUCUGC (((((..((((..........(((((((((....))))))))).................((((......))))))))..)))))................... (-24.02 = -23.13 + -0.89)

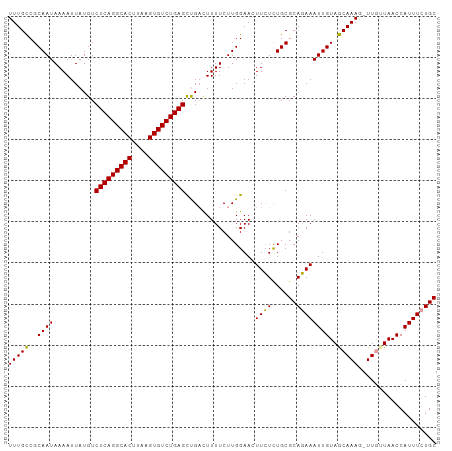
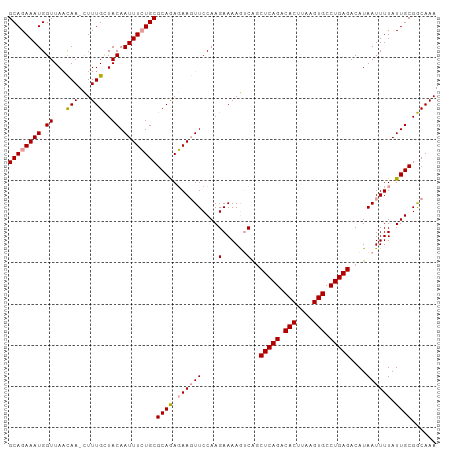
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:42 2011