| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,319,978 – 19,320,100 |
| Length | 122 |
| Max. P | 0.930578 |

| Location | 19,319,978 – 19,320,100 |
|---|---|
| Length | 122 |
| Sequences | 5 |
| Columns | 126 |
| Reading direction | reverse |
| Mean pairwise identity | 70.29 |
| Shannon entropy | 0.48920 |
| G+C content | 0.63540 |
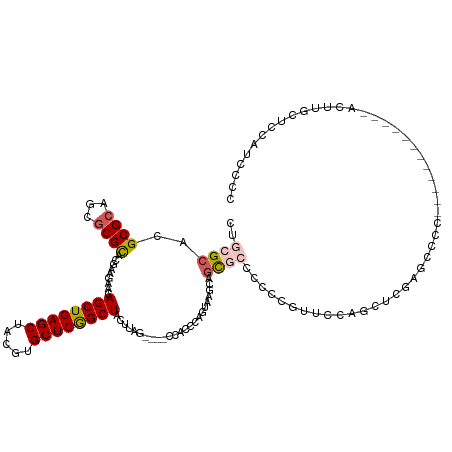
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -20.28 |
| Energy contribution | -20.92 |
| Covariance contribution | 0.64 |
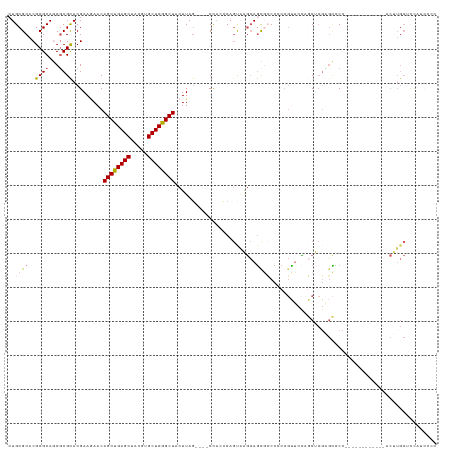
| Combinations/Pair | 1.19 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.930578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
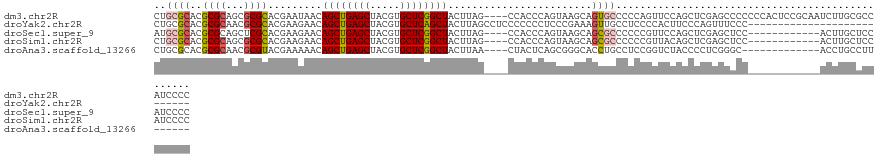
>dm3.chr2R 19319978 122 - 21146708 CUGCGCACGCGCAGCGCGCACGAAUAACAGCUGAGCUACGUGCUCGGCUACUUAG----CCACCCAGUAAGCAGUGCCCCCAGUUCCAGCUCGAGCCCCCCCACUCCGCAAUCUUGCGCCAUCCCC (((((....))))).(((((.((.....((((((((.....))))))))......----...........((((((......((((......)))).....))))..))..)).)))))....... ( -37.00, z-score = -1.91, R) >droYak2.chr2R 19286664 99 - 21139217 CUGCGCACGCGCAACGCGCACGAAGAACAGCUGAGCUACGUGCUCAGCUACUUAGCCUCCCCCCCUCCCGAAAGUUGCCUCCCCACUUCCCAGUUUCCC--------------------------- .(((((.........))))).((((...((((((((.....))))))))...................(....)...........))))..........--------------------------- ( -25.00, z-score = -2.21, R) >droSec1.super_9 2609390 110 - 3197100 AUGCGCACGCGCAGCUCGCACGAAGAACAGCUGAGCUACGUGCUCGGCUACUUAG----CCACCCAGUAAGCAGCGCCCCCCGUUCCAGCUCGAGCUCC------------ACUUGCUCCAUCCCC ....(((.(.(.((((((..(...((((((((((((.....)))))))).....(----(..........))..........))))..)..)))))).)------------.).)))......... ( -31.90, z-score = -0.69, R) >droSim1.chr2R 17900615 110 - 19596830 CUGCGCACGCGCAGCGCGCACGAAGAACAGCUGAGCUACGUGCUCGGCUACUUAG----CCACCCAGUAAGCAGCGCCCCCCGUUACAGCUCGAGCUCC------------ACUUGCUCCAUCCCC (((((....))))).((((.........((((((((.....)))))))).....(----(..........)).))))...............((((...------------....))))....... ( -33.50, z-score = -0.51, R) >droAna3.scaffold_13266 7953261 103 - 19884421 CUGCGCACGCGCAACGCGUACGAAAAACAGCUGAGCUACGUGCUCGGCUACUUAA----CUACUCAGCGGGCACCUGCCUCCGGUCUACCCCUCGGGC-------------ACCUGCCUU------ ...((((((((...))))).........((((((((.....))))))))......----.......)))((((..(((((..((......))..))))-------------)..))))..------ ( -39.10, z-score = -2.55, R) >consensus CUGCGCACGCGCAGCGCGCACGAAGAACAGCUGAGCUACGUGCUCGGCUACUUAG____CCACCCAGUAAGCAGCGCCCCCCGUUCCAGCUCGAGCCCC____________ACUUGCUCCAUCCCC ..((((..((((...)))).........((((((((.....))))))))........................))))................................................. (-20.28 = -20.92 + 0.64)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:36 2011