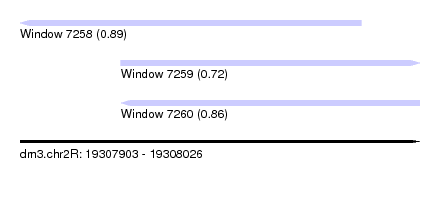
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,307,903 – 19,308,026 |
| Length | 123 |
| Max. P | 0.885381 |

| Location | 19,307,903 – 19,308,008 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 72.78 |
| Shannon entropy | 0.51004 |
| G+C content | 0.46274 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -13.86 |
| Energy contribution | -14.20 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.885381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19307903 105 - 21146708 -UCCCAACCA--UUCGGCUUUCAUUAAUUUGUUCGGCUGCAAUUGGCAAACCCCAUAUUUGCCUGCAGCUUCAUUGAGGCGUAAAGCAAAAUAAAACAGAAGAUCGAA-- -.........--((((..((((.(((.((((((..((((((...((((((.......))))))))))(((((...)))))))..)))))).)))....))))..))))-- ( -25.60, z-score = -1.36, R) >droAna3.scaffold_13266 7940070 87 - 19884421 UGGUAAACCACGCUCAGUGCUUAUUAAUUUGUCCAACUGCAAUUGGCAAUCGCC--AUUGGACGGCAGCAUCUUUGGAGCGUGAA-CGAG-------------------- ........(((((((.(((((.......((((((((.......((((....)))--))))))))).))))).....)))))))..-....-------------------- ( -26.61, z-score = -1.58, R) >droEre2.scaffold_4845 20673505 103 - 22589142 -GCCCGACCA--UUCAACUCUUAUUAACUUGCCCGGCUGCAAUUGGCAAGCCCC--AUUUGCCUGCAGCUCCGUUGAAGCGUGGUGCAAAAUGAAACCAAAAACCUAG-- -(((((....--((((((.........(......)((((((...((((((....--.))))))))))))...))))))...))).)).....................-- ( -26.00, z-score = -1.46, R) >droYak2.chr2R 19274325 105 - 21139217 -GCCCGAACA--UUCAACUCUUAUUAAUUUAUCCGGCUGCAAUUGGCAAACCCA--AUUUGCCUGCAGCUCCGUUGAAGCGUGUAGCGAAAUAAAAGCAAAUACGCCGCA -((.......--((((((...((......))...(((((((...((((((....--.)))))))))))))..))))))((((((.((.........))..)))))).)). ( -28.80, z-score = -2.32, R) >droSec1.super_9 2597777 103 - 3197100 -GACCCACAA--CUCGGCUCUUAUUAAUUUGUCCAGCUGCAAUUGGCAAAUCCC--AUUUGCCUGCAGCUUCGUUGAGGCGUAAAGCGAAAUAAAACAGAAGACCGAU-- -.........--.((((.((((.....(((((...((((((...(((((((...--)))))))))))))((((((.........)))))))))))....)))))))).-- ( -28.90, z-score = -2.47, R) >droSim1.chr2R 17888763 103 - 19596830 -GCCCCACCA--CUCGGCUCUUAUUAAUUUGUCCAACUGCAAUUGGCAAAUCCC--AUUUGCCUGCAGCUUCGUUGAGGCGUAAAGCGAAAUAAAACAGAAGAACGAA-- -.........--.(((..((((.....(((((....(((((...(((((((...--)))))))))))).((((((.........)))))))))))....)))).))).-- ( -22.10, z-score = -0.47, R) >consensus _GCCCAACCA__CUCAGCUCUUAUUAAUUUGUCCAGCUGCAAUUGGCAAACCCC__AUUUGCCUGCAGCUUCGUUGAAGCGUAAAGCGAAAUAAAACAGAAGACCGAA__ ............((((((...((......))...(((((((...((((((.......)))))))))))))..))))))((.....))....................... (-13.86 = -14.20 + 0.34)



| Location | 19,307,934 – 19,308,026 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 69.05 |
| Shannon entropy | 0.54713 |
| G+C content | 0.46139 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -13.06 |
| Energy contribution | -13.45 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.721667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19307934 92 + 21146708 UCAAUGAAGCUGCAGGCAAAUAUGGGGUUUGCCAAUUGCAGCCGAACAAAUUAAUGAAAGCCGAA--UGGUUGGGAAUUUGGC-UUUGGCCAAAG------------- ........(((((((((((((.....)))))))...))))))............(.(((((((((--(........)))))))-))).)......------------- ( -29.20, z-score = -1.76, R) >droAna3.scaffold_13266 7940082 106 + 19884421 CCAAAGAUGCUGCCGUCCAAU--GGCGAUUGCCAAUUGCAGUUGGACAAAUUAAUAAGCACUGAGCGUGGUUUACCAAAUUCGAUUAAACCGUUUCCAAAAAAAAAAG ....((.((((...(((((((--.(((((.....))))).))))))).........))))))(((..(((....)))..))).......................... ( -21.50, z-score = -0.55, R) >droEre2.scaffold_4845 20673536 78 + 22589142 UCAACGGAGCUGCAGGCAAAU--GGGGCUUGCCAAUUGCAGCCGGGCAAGUUAAUAAGAGUUGAA--UGGUCGGGCUUUUGG-------------------------- ......(((((.((......)--).))))).((((....((((.(((...(((((....))))).--..))).)))).))))-------------------------- ( -24.00, z-score = -0.61, R) >droYak2.chr2R 19274358 97 + 21139217 UCAACGGAGCUGCAGGCAAAU--UGGGUUUGCCAAUUGCAGCCGGAUAAAUUAAUAAGAGUUGAA--UGUUCGGGCUUUUGGUAUCUGGCUUCUGCCAGAG------- ((((.((.(((((((((((((--...)))))))...))))))((((((..(((((....))))).--))))))..)).))))..((((((....)))))).------- ( -33.00, z-score = -2.16, R) >droSec1.super_9 2597808 90 + 3197100 UCAACGAAGCUGCAGGCAAAU--GGGAUUUGCCAAUUGCAGCUGGACAAAUUAAUAAGAGCCGAG--UUGUGGGUCAUUUGGU-UUUGGCCAAAG------------- .......((((((((((((((--...)))))))...)))))))..............((.(((..--...))).)).((((((-....)))))).------------- ( -27.50, z-score = -1.54, R) >droSim1.chr2R 17888794 89 + 19596830 UCAACGAAGCUGCAGGCAAAU--GGGAUUUGCCAAUUGCAGUUGGACAAAUUAAUAAGAGCCGAG--UGGUGGGGCAUUUGGU-UUUGGCCAAG-------------- .......((((((((((((((--...)))))))...)))))))((...........(((((((((--((......))))))))-)))..))...-------------- ( -26.92, z-score = -1.52, R) >consensus UCAACGAAGCUGCAGGCAAAU__GGGGUUUGCCAAUUGCAGCCGGACAAAUUAAUAAGAGCCGAA__UGGUCGGGCAUUUGGU_UUUGGCCAAAG_____________ ........(((((((((((((.....)))))))...)))))).................................................................. (-13.06 = -13.45 + 0.39)


| Location | 19,307,934 – 19,308,026 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 69.05 |
| Shannon entropy | 0.54713 |
| G+C content | 0.46139 |
| Mean single sequence MFE | -22.11 |
| Consensus MFE | -11.29 |
| Energy contribution | -12.18 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.856349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19307934 92 - 21146708 -------------CUUUGGCCAAA-GCCAAAUUCCCAACCA--UUCGGCUUUCAUUAAUUUGUUCGGCUGCAAUUGGCAAACCCCAUAUUUGCCUGCAGCUUCAUUGA -------------.((((((....-))))))....(((((.--...)).................(((((((...((((((.......)))))))))))))...))). ( -22.60, z-score = -1.15, R) >droAna3.scaffold_13266 7940082 106 - 19884421 CUUUUUUUUUUGGAAACGGUUUAAUCGAAUUUGGUAAACCACGCUCAGUGCUUAUUAAUUUGUCCAACUGCAAUUGGCAAUCGCC--AUUGGACGGCAGCAUCUUUGG .........(((((...((((((.(((....)))))))))..((.....))...........)))))((((...((((....)))--).......))))......... ( -21.70, z-score = 0.07, R) >droEre2.scaffold_4845 20673536 78 - 22589142 --------------------------CCAAAAGCCCGACCA--UUCAACUCUUAUUAACUUGCCCGGCUGCAAUUGGCAAGCCCC--AUUUGCCUGCAGCUCCGUUGA --------------------------...............--.(((((.........(......)((((((...((((((....--.))))))))))))...))))) ( -17.40, z-score = -0.89, R) >droYak2.chr2R 19274358 97 - 21139217 -------CUCUGGCAGAAGCCAGAUACCAAAAGCCCGAACA--UUCAACUCUUAUUAAUUUAUCCGGCUGCAAUUGGCAAACCCA--AUUUGCCUGCAGCUCCGUUGA -------.((((((....)))))).................--.(((((...((......))...(((((((...((((((....--.)))))))))))))..))))) ( -27.40, z-score = -2.96, R) >droSec1.super_9 2597808 90 - 3197100 -------------CUUUGGCCAAA-ACCAAAUGACCCACAA--CUCGGCUCUUAUUAAUUUGUCCAGCUGCAAUUGGCAAAUCCC--AUUUGCCUGCAGCUUCGUUGA -------------.(((((.....-.))))).......(((--(..(((............))).(((((((...(((((((...--))))))))))))))..)))). ( -24.10, z-score = -2.46, R) >droSim1.chr2R 17888794 89 - 19596830 --------------CUUGGCCAAA-ACCAAAUGCCCCACCA--CUCGGCUCUUAUUAAUUUGUCCAACUGCAAUUGGCAAAUCCC--AUUUGCCUGCAGCUUCGUUGA --------------.((((.((((-.......(((......--...))).........)))).))))(((((...(((((((...--))))))))))))......... ( -19.49, z-score = -1.29, R) >consensus _____________CAUUGGCCAAA_ACCAAAUGCCCAACCA__CUCAGCUCUUAUUAAUUUGUCCAGCUGCAAUUGGCAAACCCC__AUUUGCCUGCAGCUUCGUUGA .................................................................(((((((...((((((.......)))))))))))))....... (-11.29 = -12.18 + 0.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:32 2011