
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,305,910 – 19,306,046 |
| Length | 136 |
| Max. P | 0.880737 |

| Location | 19,305,910 – 19,306,006 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 74.73 |
| Shannon entropy | 0.41528 |
| G+C content | 0.47794 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -14.52 |
| Energy contribution | -16.52 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.880737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
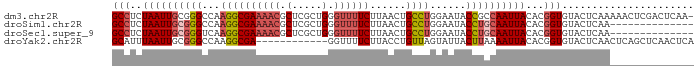
>dm3.chr2R 19305910 96 - 21146708 GCCUCUAAUUGCGGGCCAAGGCGAAAACGCUCGCUGGGUUUUCUUAACUGCCUGGAAUACCGCCAAUUACACGGUGUACUCAAAAACUCGACUCAA- (((..(((((((((.(((.(((((((((.(.....).))))))......))))))....))).))))))...))).....................- ( -24.90, z-score = -1.16, R) >droSim1.chr2R 17886822 83 - 19596830 GCCUCUAAUUGCGGGCCAAGGCGAAAACGCUCGCUGGGUUUUCUUAACUGCCUGGAAUACCUGCAAUUACACGGUGUACUCAA-------------- (((..(((((((((((((.(((((((((.(.....).))))))......))))))....))))))))))...)))........-------------- ( -27.30, z-score = -2.50, R) >droSec1.super_9 2595809 83 - 3197100 GCCUCUAAUUGCGGGUCAAGGCGAAAACGCUCGCUGGGUUUUCUUAACUGCCUGGAAUACCUGCAAUUACACGGUGUACUCAA-------------- (((..(((((((((((..((((((((((.(.....).))))))......)))).....)))))))))))...)))........-------------- ( -27.50, z-score = -2.98, R) >droYak2.chr2R 19272294 85 - 21139217 GCAUUUAAUUGCGGGCCAAGGCGA------------GGUUUUCUUACCUGUUAGUAUUACUUAAAAUUACACGGUGUACUCAACUCAGCUCAACUCA (((......)))((((..(((..(------------(.....))..)))((((((((.(((...........)))))))).)))...))))...... ( -16.60, z-score = -0.64, R) >consensus GCCUCUAAUUGCGGGCCAAGGCGAAAACGCUCGCUGGGUUUUCUUAACUGCCUGGAAUACCUGCAAUUACACGGUGUACUCAA______________ (((..((((((((((...((((((((((.(.....).))))))......))))......))))))))))...)))...................... (-14.52 = -16.52 + 2.00)

| Location | 19,305,939 – 19,306,046 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 68.56 |
| Shannon entropy | 0.52250 |
| G+C content | 0.48539 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -11.62 |
| Energy contribution | -10.98 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19305939 107 - 21146708 UCCGUCGUCCGGCAAUUAAGAAUUUUUAGUGCGCGAUUCCGCCUCUAAUUGCGGGCCAAGGCGA---------AAACGCUCGCUGGGUUUUCUUAACUGCCUGGAAUACCGCCAAU ...((..(((((((.(((((((..(((((((.(((...((((........))))((....))..---------...))).)))))))..))))))).)))).)))..))....... ( -39.30, z-score = -2.61, R) >droSim1.chr2R 17886838 107 - 19596830 CCCGUCGUCCGGCAAUUAAGAAUUUUUAGUGCGCGAUUCCGCCUCUAAUUGCGGGCCAAGGCGAAAACGCUCGCUGGGUUUUCUUAACUGCCUGGAAUACCUGCAAU--------- ...((..(((((((.(((((((..(((((((.(((...((((........))))((....)).....))).)))))))..))))))).)))).)))..)).......--------- ( -39.30, z-score = -2.79, R) >droSec1.super_9 2595825 107 - 3197100 UCCGUCGUCCGGCAAUUAAGAAUUUUUAGUGCGCGAUUCCGCCUCUAAUUGCGGGUCAAGGCGA---------AAACGCUCGCUGGGUUUUCUUAACUGCCUGGAAUACCUGCAAU ...((..(((((((.(((((((..(((((((.(((.((.(((((..............))))).---------)).))).)))))))..))))))).)))).)))..))....... ( -37.44, z-score = -2.57, R) >droYak2.chr2R 19272324 88 - 21139217 -------AACCGCAAUUAGGAAUUUUUAGUGCGCGAUUCCGCAUUUAAUUGCGGGCCAAGGCGA------------GGUUUUCUUACCUGUUAGUAUUACUUAAAAU--------- -------..((((((((((.........(((((......)))))))))))))))((....)).(------------(((......))))..................--------- ( -22.60, z-score = -1.31, R) >droEre2.scaffold_4845 20671683 85 - 22589142 ----UCGACCGGCAAUUAAGAA-UUUUAGUGCGCGAUUCCGUCUUUAAUUGCGGACCAAGGCGA---------AAACGCCCGGCGGG--------AUUACCUGCAAU--------- ----.((.(((.(((((((((.-.....(....).......)).)))))))))).....((((.---------...))))))(((((--------....)))))...--------- ( -23.22, z-score = -0.07, R) >consensus _CCGUCGUCCGGCAAUUAAGAAUUUUUAGUGCGCGAUUCCGCCUCUAAUUGCGGGCCAAGGCGA_________AAACGCUCGCUGGGCUGUCUGAACUACCUGCAAU_________ ..........(((.((((((....)))))).(((...(((((........))))).....)))..............))).................................... (-11.62 = -10.98 + -0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:29 2011