
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,252,843 – 19,252,956 |
| Length | 113 |
| Max. P | 0.568306 |

| Location | 19,252,843 – 19,252,956 |
|---|---|
| Length | 113 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 60.69 |
| Shannon entropy | 0.88435 |
| G+C content | 0.49569 |
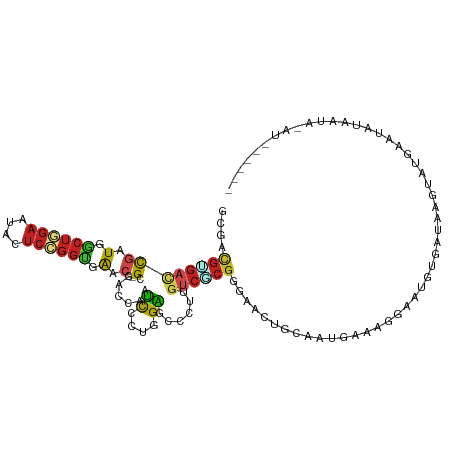
| Mean single sequence MFE | -29.01 |
| Consensus MFE | -9.32 |
| Energy contribution | -9.71 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.80 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.568306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19252843 113 + 21146708 GCGACGUGACCGAUGGCUGGAAUACUCCGGUGAAC--GCGACCAAUCCCUGGAGCCCUUGUCGCGGGAACUGCAAUGAAAGGUCUGUGAUAAGUCUGAAUAUAAUAAAUAAAACA ((..((((.....(.((((((....)))))).).)--))).(((.....))).)).(((((((((((..((........)).)))))))))))...................... ( -26.60, z-score = -0.13, R) >droSim1.chr2R 17836533 113 + 19596830 GCGACGUGACCGAUGGCUGGAAUACUCCGGUGAAC--GCGACCAAUCCCUGGAGCCCUUGUCGCGGGAACUGCAAGGAAAGGUCUGUGAUAAGUCUGAAUAUAAUAAAUAGAAAA ..(((........(.((((((....)))))).).(--((((((..(((.((.(((((.......)))..)).)).)))..)))).)))....))).................... ( -30.10, z-score = -0.82, R) >droSec1.super_9 2547281 113 + 3197100 GCGACGUGACCGAUGGCUGGAAUACUCCGGUGAAC--GCGACCAAUCCCUGGAGCCCUUGUCGCGGGAACUGCAGGGAAAGGUCUGUGAUAAGUCUGAAUAUAAUAAAUAGAAAA ..(((........(.((((((....)))))).).(--((((((..((((((.(((((.......)))..)).))))))..)))).)))....))).................... ( -36.60, z-score = -2.41, R) >droYak2.chr2R 19221555 113 + 21139217 GCGACGCGACCGAUGGCUGGAGUAUUCCGGUGAAC--GCGACCAGUCCCUGGAGCCCUUGUCGCGGGAACUGCAAUGAAUGGUUUGUGAUAAGAUUGAAUUUAAUAUAAAGAAGA ((..((((.....(.(((((......))))).).)--))).((((...)))).)).((((((((..(((((.........)))))))))))))...................... ( -27.00, z-score = 0.42, R) >droEre2.scaffold_4845 20623150 113 + 22589142 UCGACGCGACCGAUGGCUGGAGUAUUCCGGUGAAC--GCGACCAAUCCCUAGAGCCCUUGUCGCGCGAACUGCAAUGAAAGGAUUGUGAUAAGUUUGAAUUUAAUAUAUAGAAAA ....((((((.((.(((((((....((((.....)--).))....)))....)))).))))))))(((((((((((......)))))....)))))).................. ( -26.90, z-score = -0.60, R) >droAna3.scaffold_13266 9008697 110 + 19884421 CCUUCGGGAUCGAUGGCUGGAGUAUUCGGGUGAUC--GUGACCAAUCCCUCGAACCCUUGUCCCGGGAACUGGUUUGAAAGACACAGGUUUAAAAGGGAUUAUAAAGUUUUG--- .....((...((((.(((.(......).))).)))--)...))(((((((.(((((..((((((((...)))).......))))..)))))...)))))))...........--- ( -32.51, z-score = -0.24, R) >dp4.chr3 4996836 88 + 19779522 GCGACGAGACCGAUGGCUGGAAUACUCCGGUGAAC--GCGACCAAUCUCUCGAUCCCUUGUCGCGCGAACUGAAAUCAAAAUAAAGGACU------------------------- ((((((((..((.(.((((((....)))))).).)--)(((........)))....))))))))..........................------------------------- ( -23.40, z-score = -1.29, R) >droPer1.super_2 5196087 88 + 9036312 GCGACGAGACCGAUGGCUGGAAUACUCCGGUGAAC--GCGACCAAUCUCUCGAUCCCUUGUCGCGCGAACUGAAAUCAAAAUAAAAGACU------------------------- ((((((((..((.(.((((((....)))))).).)--)(((........)))....))))))))..........................------------------------- ( -23.40, z-score = -1.88, R) >droWil1.scaffold_181141 1016058 107 - 5303230 GCGACGUGAUCGGUGACUUGAAUACUCGGGUGAAC--GUGACCAAUCACGAGGGGGCUUCUCCCUAGGACUGCAAAUACAAUACUGUACAACUGAAAAAAUCAAAAACU------ (((.((((((.(((.((((((....))))(....)--)).))).))))))((((((...)))))).....)))...((((....)))).....................------ ( -25.10, z-score = -1.20, R) >droVir3.scaffold_12875 2196633 101 - 20611582 GCGACGUGAGCGGUGGCUCGAAUACUCUGGCGAUC--GCGACCAAUCACGAGGCGCCUUGUCACGAGAACUGUACAGAGAAAGAUUAGAGAAUGAAUAGAACA------------ ....((((((((((.(((.((....)).))).)))--)).........((((....)))))))))....((((.((................)).))))....------------ ( -24.29, z-score = 0.51, R) >droMoj3.scaffold_6496 14801733 111 - 26866924 GCGUCGCGCUCUGUGACUGGAAUACUCCGGUGAGC--GCGACCAAUCACGCGGCGCCUUGUCGCGUGAGCUGUUAAUG-GAUAUUUUAAAUUAUGAAAAAUUGAAAGUGGAGUA- (((((((((((....((((((....))))))))))--)))))...(((((((((.....)))))))))))........-..((((((.((((......)))).)))))).....- ( -42.60, z-score = -3.63, R) >anoGam1.chr3R 3105054 98 - 53272125 GCUUUGUACUUCGUUCAGAGUGUGCUGCAGCUGCUUGGCGAUCUUCUCCUGGGCGGCCUGU---GUACAUCACGAUCAAAGAAACGGGUUAGCAAGUGGCA-------------- .(((((....((((...((.(((((.(((((((((..(.((.....)))..))))).))))---))))))))))).)))))..........((.....)).-------------- ( -29.60, z-score = 0.44, R) >consensus GCGACGUGACCGAUGGCUGGAAUACUCCGGUGAAC__GCGACCAAUCCCUGGAGCCCUUGUCGCGGGAACUGCAAUGAAAGGAAUGUGAUAAGUAUGAAUAUAAUA_AU______ ....((((((...(.((((((....)))))).)............((....))......)))))).................................................. ( -9.32 = -9.71 + 0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:25 2011