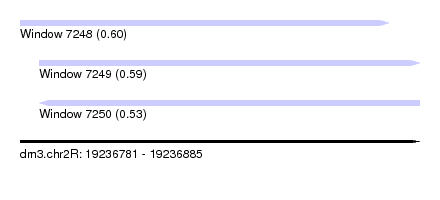
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,236,781 – 19,236,885 |
| Length | 104 |
| Max. P | 0.599082 |

| Location | 19,236,781 – 19,236,877 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 64.97 |
| Shannon entropy | 0.70395 |
| G+C content | 0.43647 |
| Mean single sequence MFE | -25.74 |
| Consensus MFE | -7.40 |
| Energy contribution | -7.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.599082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
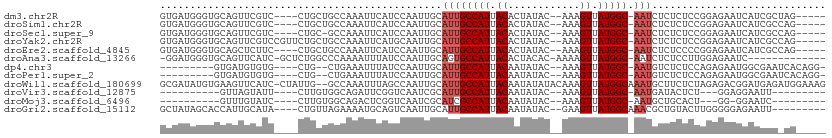
>dm3.chr2R 19236781 96 + 21146708 --------CUCCGCUAGCGAUGAUUCUCCGGAGAGAGAUU-GCCAUAACUUU--GUAUAGUGUAAUGGCAAUGCAAUUGGAUGAAUUU-GGCAGCAG----GACGAACUGCA --------....((((.....((((((((((...(..(((-(((((.(((..--....)))...)))))))).)..))))).))))))-))).((((----......)))). ( -27.70, z-score = -1.36, R) >droSim1.chr2R 17820064 96 + 19596830 --------CUCCGCUGGCGAUGAUUCUCCGGAGAGAGAUU-GCCAUAACUUU--GUAUAGUGUAAUGGCAAUGCAAUUGGAUGAAUUU-GGCAGCAG----GACGAACUGCA --------.(((((((.(((..(((((((((...(..(((-(((((.(((..--....)))...)))))))).)..))))).))))))-).)))).)----))......... ( -29.40, z-score = -1.62, R) >droSec1.super_9 2529845 95 + 3197100 --------CUCCGCUGGCGAUGAUUCUCCGGAGAGAGAUU-GCCAUAACUUU--GUAUAGUGUAAUGGCAAUGCAAUUGGAUGAAUUU-GGC-GCAG----GACGAACUGCA --------((((.((((.((....)).)))).).)))(((-(((((.(((..--....)))...))))))))................-...-((((----......)))). ( -27.80, z-score = -1.21, R) >droYak2.chr2R 19203162 100 + 21139217 --------CUCCGCUGGCGAUGAUUCUCCGGAGAGAGAUU-GCCAUAACUUU--GUAUAGUGUAAUGGCAAUGCAAUUGCAUGAAUUU-GGCAGCAGAACGGACGAACUGCA --------((((.((((.((....)).)))).).)))(((-(((((.(((..--....)))...))))))))((....))........-....((((..(....)..)))). ( -26.90, z-score = -0.47, R) >droEre2.scaffold_4845 20607144 96 + 22589142 --------CUCCGCUGGCGAUGAUUCUCCGGGGAGAGAUU-GCCAUAACUUU--GUAUAGUGUAAUGGCAAUGCAAUUGGAUGAAUUU-GGCAGCAG----GAAGAGCUGCA --------((((.((((.((....)).))))))))..(((-(((((.(((..--....)))...))))))))................-.(((((..----.....))))). ( -32.70, z-score = -1.96, R) >droAna3.scaffold_13266 8330411 87 - 19884421 ---------------------GAUUCUCCAAGGAGAGAUU-GCCAUAACUUUU-GUGUAGUGUAAUGGCACUGCAAUUGGAUAAAUUU-UGGGCCAGAGC-GAUGAACUGCA ---------------------..(((((....)))))(((-((..........-.((((((((....)))))))).((((........-....)))).))-)))........ ( -20.80, z-score = -0.29, R) >dp4.chr3 16748096 89 - 19779522 ----UUCCGCCUGUGAUUCGCCAUUCUCUGGAGAGACAUU-GCCAUAACUUU--GUAUAUUGUAAUGGCAAUGCAAUUGGAUAAAUUUCAGCAGCA---------------- ----....(((((.((((..(((.((((....))))((((-(((((.((...--.......)).)))))))))....)))...)))).)))..)).---------------- ( -24.40, z-score = -1.69, R) >droPer1.super_2 5811197 89 - 9036312 ----UUCCGCCUGUGAUUCGCCAUUCUCUGGAGAGACAUU-GCCAUAACUUU--GUAUAUUGUAAUGGCAAUGCAAUUGGAUAAAUUUCAGCAGCA---------------- ----....(((((.((((..(((.((((....))))((((-(((((.((...--.......)).)))))))))....)))...)))).)))..)).---------------- ( -24.40, z-score = -1.69, R) >droWil1.scaffold_180699 117196 108 + 2593675 CUUUCUCUCUUUCCAUCUCAUCCGUCUCUAGAGAAGCAUUUGCCAUAACUUUGUAUAUAUUGUAAUGGCAAUGCAAUUGGCUAAAUUUGGCCAAUAG----GAUGAACUUCA .................((((((.(((....))).(((((.(((((.((..((....))..)).)))))))))).((((((((....)))))))).)----)))))...... ( -32.00, z-score = -4.35, R) >droVir3.scaffold_12875 3251766 83 + 20611582 ------------CUCGGAAUUCCUCC---AGAGUAUCAUU-GCCAUAACUUU--GUAUAUUGUAAUGGCAAUGCGAUUGACCGAAUCU-GCCACAAGAAUAC---------- ------------.((((.((((....---.))))..((((-(((((.((...--.......)).))))))))).......)))).(((-......)))....---------- ( -19.50, z-score = -2.02, R) >droMoj3.scaffold_6496 4971362 82 - 26866924 ------------CUCUGGAUUCC-CC---AGUGCAGCAUU-GCCAUAACUUU--GUAUAUUGUAAUGGCGAUGCGAUUGACCGAGUCU-GCCACAAGGAUAC---------- ------------....((((((.-.(---(((...(((((-(((((.((...--.......)).)))))))))).))))...))))))-.((....))....---------- ( -24.50, z-score = -2.15, R) >droGri2.scaffold_15112 2279867 93 - 5172618 ------------CUCUGAAUUCUCCCCCAAGUACAGCGUUUGCCAUAACUUC--GUAUAUUGUAAUGGCAAUGCAAUUGACUGCAUUU-UCUAACAGUAUGCAAUGGU---- ------------..............(((......(((((.(((((.((...--.......)).)))))))))).......(((((..-.........))))).))).---- ( -18.80, z-score = -0.66, R) >consensus ________CUCCGCUGGCGAUCAUUCUCCGGAGAGACAUU_GCCAUAACUUU__GUAUAGUGUAAUGGCAAUGCAAUUGGAUGAAUUU_GGCAGCAG____GA_GAACUGCA .........................................(((((.((............)).)))))........................................... ( -7.40 = -7.40 + -0.00)

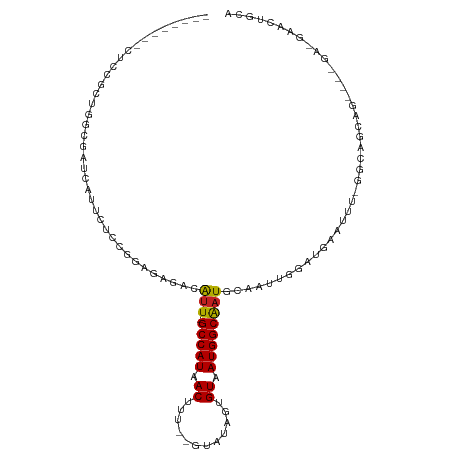
| Location | 19,236,786 – 19,236,885 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 66.20 |
| Shannon entropy | 0.69679 |
| G+C content | 0.43257 |
| Mean single sequence MFE | -25.33 |
| Consensus MFE | -7.40 |
| Energy contribution | -7.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.593750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19236786 99 + 21146708 -----CUAGCGAUGAUUCUCCGGAGAGAGAUU-GCCAUAACUUU--GUAUAGUGUAAUGGCAAUGCAAUUGGAUGAAUUUGGCAGCAG----GACGAACUGCACCCAUCAC -----.....(((((((((((((...(..(((-(((((.(((..--....)))...)))))))).)..))))).))))..((..((((----......)))).)))))).. ( -27.40, z-score = -1.55, R) >droSim1.chr2R 17820069 99 + 19596830 -----CUGGCGAUGAUUCUCCGGAGAGAGAUU-GCCAUAACUUU--GUAUAGUGUAAUGGCAAUGCAAUUGGAUGAAUUUGGCAGCAG----GACGAACUGCACCCAUCAC -----.....(((((((((((((...(..(((-(((((.(((..--....)))...)))))))).)..))))).))))..((..((((----......)))).)))))).. ( -27.40, z-score = -1.21, R) >droSec1.super_9 2529850 98 + 3197100 -----CUGGCGAUGAUUCUCCGGAGAGAGAUU-GCCAUAACUUU--GUAUAGUGUAAUGGCAAUGCAAUUGGAUGAAUUUGGC-GCAG----GACGAACUGCACCCAUCAC -----.....(((((((((((((...(..(((-(((((.(((..--....)))...)))))))).)..))))).))))..((.-((((----......)))).)))))).. ( -29.90, z-score = -2.14, R) >droYak2.chr2R 19203167 103 + 21139217 -----CUGGCGAUGAUUCUCCGGAGAGAGAUU-GCCAUAACUUU--GUAUAGUGUAAUGGCAAUGCAAUUGCAUGAAUUUGGCAGCAGAACGGACGAACUGCACCCAUCAC -----...(((((..(((((....)))))(((-(((((.(((..--....)))...))))))))...))))).(((...(((..((((..(....)..))))..)))))). ( -26.40, z-score = -0.73, R) >droEre2.scaffold_4845 20607149 99 + 22589142 -----CUGGCGAUGAUUCUCCGGGGAGAGAUU-GCCAUAACUUU--GUAUAGUGUAAUGGCAAUGCAAUUGGAUGAAUUUGGCAGCAG----GAAGAGCUGCACCCAUCAC -----.....(((((((((((((...(..(((-(((((.(((..--....)))...)))))))).)..))))).))))...(((((..----.....)))))...)))).. ( -30.20, z-score = -1.34, R) >droAna3.scaffold_13266 8330411 94 - 19884421 -------------GAUUCUCCAAGGAGAGAUU-GCCAUAACUUUU-GUGUAGUGUAAUGGCACUGCAAUUGGAUAAAUUUUGGGCCAGAGC-GAUGAACUGCACCCAUCC- -------------..(((((....)))))...-............-(((((((...((.((.(((...(..((.....))..)..))).))-.))..)))))))......- ( -23.90, z-score = -0.69, R) >dp4.chr3 16748101 92 - 19779522 -CCUGUGAUUCGCCAUUCUCUGGAGAGACAUU-GCCAUAACUUU--GUAUAUUGUAAUGGCAAUGCAAUUGGAUAAAUUUCAG--CAG----CACACAUCAC--------- -.(((.((((..(((.((((....))))((((-(((((.((...--.......)).)))))))))....)))...)))).)))--...----..........--------- ( -22.50, z-score = -1.24, R) >droPer1.super_2 5811202 92 - 9036312 -CCUGUGAUUCGCCAUUCUCUGGAGAGACAUU-GCCAUAACUUU--GUAUAUUGUAAUGGCAAUGCAAUUGGAUAAAUUUCAG--CAG----CACACAUCAC--------- -.(((.((((..(((.((((....))))((((-(((((.((...--.......)).)))))))))....)))...)))).)))--...----..........--------- ( -22.50, z-score = -1.24, R) >droWil1.scaffold_180699 117204 108 + 2593675 CUUUCCAUCUCAUCCGUCUCUAGAGAAGCAUUUGCCAUAACUUUGUAUAUAUUGUAAUGGCAAUGCAAUUGGCUAAAUUUGGC--CAAUAG-GAUGAACUUCACAUAUCGC .........((((((.(((....))).(((((.(((((.((..((....))..)).)))))))))).((((((((....))))--)))).)-))))).............. ( -32.00, z-score = -3.94, R) >droVir3.scaffold_12875 3251771 82 + 20611582 ---------AAUUCCUCC---AGAGUAUCAUU-GCCAUAACUUU--GUAUAUUGUAAUGGCAAUGCGAUUGACCGAAUCUGCCACAAG----AAUACUAAC---------- ---------.........---..(((((((((-(((((.((...--.......)).)))))))))............(((......))----))))))...---------- ( -16.30, z-score = -1.93, R) >droMoj3.scaffold_6496 4971367 81 - 26866924 ---------GAUUCC-CC---AGUGCAGCAUU-GCCAUAACUUU--GUAUAUUGUAAUGGCGAUGCGAUUGACCGAGUCUGCCACAAG----GAUACAAAC---------- ---------...(((-..---.((((((((((-(((((.((...--.......)).))))))))))((((.....)))))).)))..)----)).......---------- ( -23.60, z-score = -2.73, R) >droGri2.scaffold_15112 2279872 96 - 5172618 ---------AAUUCUCCCCCAAGUACAGCGUUUGCCAUAACUUC--GUAUAUUGUAAUGGCAAUGCAAUUGACUGCAUUUUCUAACAG----UAUGCAAUGGUGCUAUAGC ---------............(((((.(((((.(((((.((...--.......)).)))))))))).......(((((..........----.)))))...)))))..... ( -21.90, z-score = -0.96, R) >consensus _____CU_GCGAUCAUUCUCCGGAGAGACAUU_GCCAUAACUUU__GUAUAGUGUAAUGGCAAUGCAAUUGGAUGAAUUUGGCAGCAG____GACGAACUGCACCCAUCAC .................................(((((.((............)).))))).................................................. ( -7.40 = -7.40 + -0.00)



| Location | 19,236,786 – 19,236,885 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 66.20 |
| Shannon entropy | 0.69679 |
| G+C content | 0.43257 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -5.41 |
| Energy contribution | -5.41 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.532698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19236786 99 - 21146708 GUGAUGGGUGCAGUUCGUC----CUGCUGCCAAAUUCAUCCAAUUGCAUUGCCAUUACACUAUAC--AAAGUUAUGGC-AAUCUCUCUCCGGAGAAUCAUCGCUAG----- (((((((((((((......----)))).)))..((((.(((......((((((((...(((....--..))).)))))-)))........)))))))))))))...----- ( -29.54, z-score = -3.00, R) >droSim1.chr2R 17820069 99 - 19596830 GUGAUGGGUGCAGUUCGUC----CUGCUGCCAAAUUCAUCCAAUUGCAUUGCCAUUACACUAUAC--AAAGUUAUGGC-AAUCUCUCUCCGGAGAAUCAUCGCCAG----- (((((((((((((......----)))).)))..((((.(((......((((((((...(((....--..))).)))))-)))........)))))))))))))...----- ( -29.14, z-score = -2.80, R) >droSec1.super_9 2529850 98 - 3197100 GUGAUGGGUGCAGUUCGUC----CUGC-GCCAAAUUCAUCCAAUUGCAUUGCCAUUACACUAUAC--AAAGUUAUGGC-AAUCUCUCUCCGGAGAAUCAUCGCCAG----- (((((((((((((......----))))-)))..((((.(((......((((((((...(((....--..))).)))))-)))........)))))))))))))...----- ( -32.94, z-score = -4.30, R) >droYak2.chr2R 19203167 103 - 21139217 GUGAUGGGUGCAGUUCGUCCGUUCUGCUGCCAAAUUCAUGCAAUUGCAUUGCCAUUACACUAUAC--AAAGUUAUGGC-AAUCUCUCUCCGGAGAAUCAUCGCCAG----- (((((((((((((..(....)..)))).))).......(((....)))(((((((...(((....--..))).)))))-))(((((....)))))..))))))...----- ( -31.40, z-score = -2.78, R) >droEre2.scaffold_4845 20607149 99 - 22589142 GUGAUGGGUGCAGCUCUUC----CUGCUGCCAAAUUCAUCCAAUUGCAUUGCCAUUACACUAUAC--AAAGUUAUGGC-AAUCUCUCCCCGGAGAAUCAUCGCCAG----- ((((((((.(((((.....----..))))))................((((((((...(((....--..))).)))))-))).((((....)))).)))))))...----- ( -30.20, z-score = -3.11, R) >droAna3.scaffold_13266 8330411 94 + 19884421 -GGAUGGGUGCAGUUCAUC-GCUCUGGCCCAAAAUUUAUCCAAUUGCAGUGCCAUUACACUACAC-AAAAGUUAUGGC-AAUCUCUCCUUGGAGAAUC------------- -((((((((.(((......-...)))))))).......)))........((((((...(((....-...))).)))))-)...((((....))))...------------- ( -22.61, z-score = -0.67, R) >dp4.chr3 16748101 92 + 19779522 ---------GUGAUGUGUG----CUG--CUGAAAUUUAUCCAAUUGCAUUGCCAUUACAAUAUAC--AAAGUUAUGGC-AAUGUCUCUCCAGAGAAUGGCGAAUCACAGG- ---------(((((...((----((.--..................(((((((((.((.......--...)).)))))-))))((((....))))..)))).)))))...- ( -26.10, z-score = -2.02, R) >droPer1.super_2 5811202 92 + 9036312 ---------GUGAUGUGUG----CUG--CUGAAAUUUAUCCAAUUGCAUUGCCAUUACAAUAUAC--AAAGUUAUGGC-AAUGUCUCUCCAGAGAAUGGCGAAUCACAGG- ---------(((((...((----((.--..................(((((((((.((.......--...)).)))))-))))((((....))))..)))).)))))...- ( -26.10, z-score = -2.02, R) >droWil1.scaffold_180699 117204 108 - 2593675 GCGAUAUGUGAAGUUCAUC-CUAUUG--GCCAAAUUUAGCCAAUUGCAUUGCCAUUACAAUAUAUACAAAGUUAUGGCAAAUGCUUCUCUAGAGACGGAUGAGAUGGAAAG .............((((((-(.((((--((........)))))).((((((((((.((............)).))))).)))))............)))))))........ ( -27.40, z-score = -1.89, R) >droVir3.scaffold_12875 3251771 82 - 20611582 ----------GUUAGUAUU----CUUGUGGCAGAUUCGGUCAAUCGCAUUGCCAUUACAAUAUAC--AAAGUUAUGGC-AAUGAUACUCU---GGAGGAAUU--------- ----------......(((----(((.(((.((...(((....)))(((((((((.((.......--...)).)))))-))))...))))---).)))))).--------- ( -18.70, z-score = -1.19, R) >droMoj3.scaffold_6496 4971367 81 + 26866924 ----------GUUUGUAUC----CUUGUGGCAGACUCGGUCAAUCGCAUCGCCAUUACAAUAUAC--AAAGUUAUGGC-AAUGCUGCACU---GG-GGAAUC--------- ----------.......((----(((...((((((...)))....((((.(((((.((.......--...)).)))))-.)))))))...---))-)))...--------- ( -21.70, z-score = -1.26, R) >droGri2.scaffold_15112 2279872 96 + 5172618 GCUAUAGCACCAUUGCAUA----CUGUUAGAAAAUGCAGUCAAUUGCAUUGCCAUUACAAUAUAC--GAAGUUAUGGCAAACGCUGUACUUGGGGGAGAAUU--------- ..((((((.....((((.(----((((........)))))....))))(((((((.((.......--...)).)))))))..))))))..............--------- ( -23.60, z-score = -0.96, R) >consensus GUGAUGGGUGCAGUUCGUC____CUGCUGCCAAAUUCAUCCAAUUGCAUUGCCAUUACAAUAUAC__AAAGUUAUGGC_AAUCUCUCUCCGGAGAAUCAUCGC_AG_____ ..................................................(((((.((............)).)))))................................. ( -5.41 = -5.41 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:23 2011