| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,183,117 – 19,183,227 |
| Length | 110 |
| Max. P | 0.948465 |

| Location | 19,183,117 – 19,183,227 |
|---|---|
| Length | 110 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.16 |
| Shannon entropy | 0.38798 |
| G+C content | 0.40981 |
| Mean single sequence MFE | -28.49 |
| Consensus MFE | -13.33 |
| Energy contribution | -13.34 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
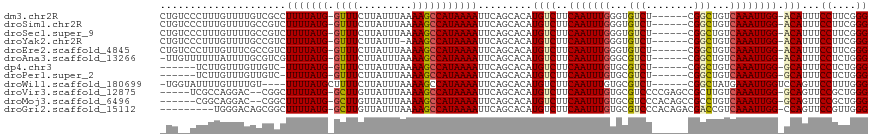
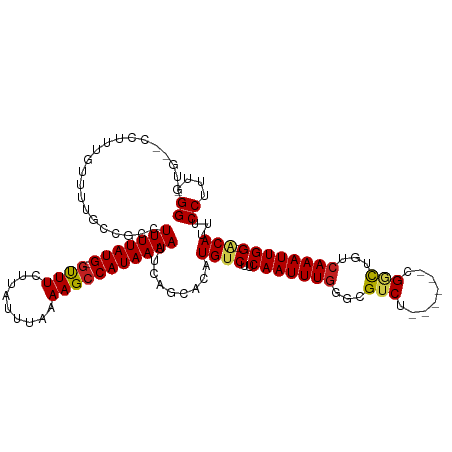
>dm3.chr2R 19183117 110 - 21146708 CUGUCCCUUUGUUUUGUCGCCUUUUAUG-GUUUCUUAUUUAAAAGCCAUAAAAUUCAGCACAUGUCUUCAAUUUGGGUGUCU------CGGCUGUCAAAUUGG-ACAUUUCCUUCGGG ....(((.......(((.(..(((((((-((((.........)))))))))))..).))).(((((..((((((((..((..------..))..)))))))))-)))).......))) ( -25.50, z-score = -2.23, R) >droSim1.chr2R 17766705 110 - 19596830 CUGUCCCUUUGUUUUGCCGUCUUUUAUG-GUUUCUUAUUUAAAAGCCAUAAAAUUCAGCACAUGUCUUCAAUUUGGGUGUCU------CGGCUGUCAAAUUGG-ACAUUUCCUUCGGG ....(((.......(((.(..(((((((-((((.........)))))))))))..).))).(((((..((((((((..((..------..))..)))))))))-)))).......))) ( -27.90, z-score = -3.09, R) >droSec1.super_9 2476240 110 - 3197100 CUGUCCCUUUGUUUUGCCGUCUUUUAUG-GUUUCUUAUUUAAAAGCCAUAAAAUUCAGCACAUGUCUUCAAUUUGGGUGUCU------CGGCUGUCAAAUUGG-ACAUUUCCUUCGGG ....(((.......(((.(..(((((((-((((.........)))))))))))..).))).(((((..((((((((..((..------..))..)))))))))-)))).......))) ( -27.90, z-score = -3.09, R) >droYak2.chr2R 19147772 109 - 21139217 CUGUCCCUUUGUUUUGCCGUCUUUUAUG-GUUUCUUAUUU-AAAGCCAUAAAAUUCAGCACAUGUCUUCAAUUUGGGUGUCU------CGGCUGUCAAAUUGG-ACAUUUCCUUCGGG ....(((.......(((.(..(((((((-((((.......-.)))))))))))..).))).(((((..((((((((..((..------..))..)))))))))-)))).......))) ( -28.70, z-score = -3.42, R) >droEre2.scaffold_4845 20550358 110 - 22589142 CUGUCCCUUUGUUUCGCCGUCUUUUAUG-GUUUCUUAUUUAAAAGCCAUAAAAUUCAGCACAUGUCUUCAAUUUGGGUGUCU------CGGCUGUCAAAUUGG-ACAUUUCCUUCGGG ....(((........((.(..(((((((-((((.........)))))))))))..).))..(((((..((((((((..((..------..))..)))))))))-)))).......))) ( -27.60, z-score = -3.11, R) >droAna3.scaffold_13266 8278558 109 + 19884421 -UUGUUUUUUAUUUUGCGUCGUUUUAUG-GUUUCUUAUUUAAAAGCCAUAAAAUUCAGCACAUGUCUUCAAUUUGGGCGUCU------CGGCUGUCAAAUUGG-ACAUUUCCUCUGGG -.............(((...((((((((-((((.........))))))))))))...))).(((((..((((((((((....------..)))..))))))))-)))).......... ( -25.10, z-score = -2.58, R) >dp4.chr3 16683906 103 + 19779522 ------UCUUGUUUGUUGUC-UUUUAUG-GUUUCUUAUUUAAAAGCCAUAAAAUUCAGCACAUGUCUUCAAUUUGUGCGUCU------CGGCUGUCAAAUUGG-GCAUUUCCUCUGGG ------.......(((((..-(((((((-((((.........)))))))))))..))))).(((((..(((((((.((....------..))...))))))))-)))).......... ( -25.60, z-score = -3.49, R) >droPer1.super_2 5747899 103 + 9036312 ------UCUUGUUUGUUGUC-UUUUAUG-GUUUCUUAUUUAAAAGCCAUAAAAUUCAGCACAUGUCUUCAAUUUGUGCGUCU------CGGCUGUCAAAUUGG-GCAUUUCCUCUGGG ------.......(((((..-(((((((-((((.........)))))))))))..))))).(((((..(((((((.((....------..))...))))))))-)))).......... ( -25.60, z-score = -3.49, R) >droWil1.scaffold_180699 188846 107 - 2593675 -UGGUAUUUUGUUUUGU----UUUUAUGCUUUUCUUAUUUAAAAGCCAUAAAAUUCAGCACAUGUCUUCAAUUUGUGCGUCU------CGGCUAUGAAAUUGGUCCAGUUCCUUUGGG -.((....(((...((.----((((((((((((.......))))).)))))))..))(((((...........)))))....------.(((((......))))))))..))...... ( -17.20, z-score = 0.08, R) >droVir3.scaffold_12875 3183340 109 - 20611582 -----UCGCCAGGAC--CGGCUUUUAUG-GCUUGUUAUUUAAAAGCCAUAAAAUUCAGCACAUGUCUUCAAUUUGUGCGUCCCCGAGCCGCUUGUCAAAUUGG-GCAGUUCCGCUGGG -----...(((((((--.(..(((((((-((((.........)))))))))))..).(((((...........)))))))))..((((.(((((......)))-)).))))...))). ( -35.70, z-score = -2.22, R) >droMoj3.scaffold_6496 4885256 108 + 26866924 ------CGGCAGGAC--CGGCUUUUAUG-GCUUGUUAUUUAAAAGCCAUAAAAUUCAGCACAUGUCUUCAAUUUGUGCGUCCCACAGCCGCCUGUCAAAUUGG-GCAGUUCCGCUGGG ------((((.((((--.((((((((((-((((.........)))))))))).....(((((...........))))).......))))(((((......)))-)).)))).)))).. ( -39.20, z-score = -3.17, R) >droGri2.scaffold_15112 2968198 107 - 5172618 ---------UGGGACAGCGGCUUUUAUG-GCUUGUUAUUUAAAAGCCAUAAAAUUCAGCACAUGUCUUCAAUUUGUGCGUCCCACAGACGACCGUCAAAUUGG-CCAGUUCCGUUGGG ---------((((((...(..(((((((-((((.........)))))))))))..).(((((...........)))))))))))..(((....))).......-((((.....)))). ( -35.90, z-score = -3.11, R) >consensus _UG__CCUUUGUUUUGCCGCCUUUUAUG_GUUUCUUAUUUAAAAGCCAUAAAAUUCAGCACAUGUCUUCAAUUUGGGCGUCU______CGGCUGUCAAAUUGG_ACAUUUCCUUUGGG .....................(((((((.((((.........))))))))))).........(((...(((((((...(((........)))...)))))))..)))...((....)) (-13.33 = -13.34 + 0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:18 2011