| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,145,155 – 19,145,247 |
| Length | 92 |
| Max. P | 0.986440 |

| Location | 19,145,155 – 19,145,247 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 64.48 |
| Shannon entropy | 0.60173 |
| G+C content | 0.46813 |
| Mean single sequence MFE | -37.82 |
| Consensus MFE | -7.34 |
| Energy contribution | -9.63 |
| Covariance contribution | 2.29 |
| Combinations/Pair | 1.50 |
| Mean z-score | -3.82 |
| Structure conservation index | 0.19 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
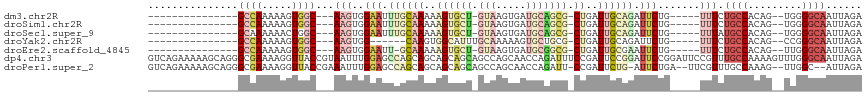
>dm3.chr2R 19145155 92 + 21146708 ---------------GCCAAAAAGUGGC---AAGUGGAAUUUGCAAAAAGUGCU-GUAAGUGAUGCAGCG-CUGAUUGCAGAUUCUG-----UUUCUGCCACAG--UGGGGCAAUUAGA ---------------(((..(..(((((---(((..((((((((((..((((((-(((.....)))))))-))..))))))))))..-----))..))))))..--)..)))....... ( -45.50, z-score = -6.75, R) >droSim1.chr2R 17727069 92 + 19596830 ---------------GCCAAAAAGUGGC---AAGUGGAAUUUGCAAAAAGUGCU-GUAAGUGAUGCAGCG-CUGAUUGCAGAUUCUG-----UUUCUGCCACAG--UGGGGCAAUUAGA ---------------(((..(..(((((---(((..((((((((((..((((((-(((.....)))))))-))..))))))))))..-----))..))))))..--)..)))....... ( -45.50, z-score = -6.75, R) >droSec1.super_9 2438580 92 + 3197100 ---------------GCAAAAAACUGGC---AAGUGGAAUUUGCAAAAAGUGCU-GUAAGUGAUGCAGCG-CUGAUUGCAGAUUCUG-----UUUAUGCCACAG--UGGGGCAAUUAGA ---------------((.......((((---(((..((((((((((..((((((-(((.....)))))))-))..))))))))))..-----))..)))))...--....))....... ( -38.34, z-score = -4.87, R) >droYak2.chr2R 19110217 87 + 21139217 ---------------GCCAAAAAGUGGC---AAGUGG------CAAGUGGCAUUUGCAAAAAGUGCUGCG-CUGAUUGCAGAUUCUG-----UUUCUGCCACAG--CCGGGCAAUUAGA ---------------(((......((((---..((((------((((..(.((((((((..((((...))-))..)))))))).)..-----))..)))))).)--))))))....... ( -31.20, z-score = -1.09, R) >droEre2.scaffold_4845 20513422 91 + 22589142 ---------------GCCAAAAAGUGGC---AAGUGGAAUU-GCAAAAAGUGCU-GUAAGUGAUGCGGCG-CUGAUUGCGAAUUCUG-----UUUCUGCCACAG--UUGGGCAAUUAGA ---------------(((.((..(((((---(((..(((((-((((..((((((-(((.....)))))))-))..)))).)))))..-----))..))))))..--)).)))....... ( -39.80, z-score = -5.13, R) >dp4.chr3 16640647 119 - 19779522 GUCAGAAAAAGCAGGGCGAAAAGGUUACCGUAAUUUGGAGCCAGCAGCAGCAGCAGCCAGCAACCAGAUUUCCGAUUCCGGAUUCCGGAUUCCGUUUGCCAAAAGUUUGGGCAAUUAGA (((...(((.....((((((..((...(((.(((((((.((..((.((....)).))..))..)))))))((((....))))...)))...)).)))))).....))).)))....... ( -31.90, z-score = -0.60, R) >droPer1.super_2 5703737 111 - 9036312 GUCAGAAAAAGCAGGGCGAAAAGGUUACCGAAAUUUGGAGCCAGCAGCAGCAGCAGCCAGCAACCAGAUU-CCGAUUCUG-AUUCUGA--UUCGUUUGCCAAAG--UUGGC--AUUAGA (((((((...((..(((.....((((.((((...)))))))).((....))....))).))...((((..-.....))))-.))))))--)((...(((((...--.))))--)...)) ( -32.50, z-score = -1.59, R) >consensus _______________GCCAAAAAGUGGC___AAGUGGAAUUUGCAAAAAGUGCU_GUAAGUGAUGCAGCG_CUGAUUGCAGAUUCUG_____UUUCUGCCACAG__UGGGGCAAUUAGA ..............(((((.....)))))......((((.((((((..((((((.(((.....))))))).))..)))))).))))..........((((.........))))...... ( -7.34 = -9.63 + 2.29)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:13 2011