
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,133,205 – 19,133,317 |
| Length | 112 |
| Max. P | 0.640773 |

| Location | 19,133,205 – 19,133,317 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 128 |
| Reading direction | reverse |
| Mean pairwise identity | 66.87 |
| Shannon entropy | 0.60136 |
| G+C content | 0.43202 |
| Mean single sequence MFE | -29.95 |
| Consensus MFE | -9.23 |
| Energy contribution | -7.10 |
| Covariance contribution | -2.13 |
| Combinations/Pair | 1.78 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.640773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19133205 112 - 21146708 -------CGAUAUCGGGCCAAAGCA-GAGAGAGAACAGCCUCUGU-----CAAAAGCCA-AUUGGCCAUUGGUAAUUGGAAUAG--GCCUGACAUCCAAUGGUGAUUAAAGUUAAUUGAACUAACACC -------.(((.(((((((......-((.((((......)))).)-----).....(((-((((.((...)))))))))....)--)))))).)))....((((.....((((.....))))..)))) ( -33.80, z-score = -2.96, R) >droEre2.scaffold_4845 20502280 116 - 22589142 -----CACGAUA-CGGGCCAAAACG---GAAAGAACAGCCUCUGCUCUGACAAAAGCCA-AGUUGUAAUUGGUAAUUGGAAAAA--ACCUGACAGCCAAUGGUGAUUUAAGUUAAUUGAACUAACACC -----.(((((.-..(((.....((---((.(((......)))..))))......))).-.))))).((((((..(..(.....--..)..)..))))))((((.....((((.....))))..)))) ( -22.30, z-score = -0.11, R) >droYak2.chr2R 19098454 96 - 21139217 -----------------------CA-GAGAGGGAACAGCCUCUGU-----CAAAAGCCA-AGUUGUAAUUGGUAAUUGGAAAAA--ACCCGACAGCCAAUGAUGAUUUAAGUUAAUUGAACUAACACC -----------------------..-.(((((......))))).(-----(((.(((.(-(((..(.((((((..((((.....--..))))..)))))).)..))))..)))..))))......... ( -21.20, z-score = -1.69, R) >droSec1.super_9 2426874 110 - 3197100 -------CGAUAUCGGGCCAAAACA---GAGGGAACAGCCUCUGU-----CAAAAGCCA-AUUGGCCAUUGGUAAUUGGAAAAA--GCCUGACAGCCAAUGCUGAUUAAAGUUAAUUGAACUAACACC -------.....((((((....(((---((((......)))))))-----......(((-((((.((...))))))))).....--))))))((((....))))......((((.......))))... ( -31.60, z-score = -2.53, R) >droSim1.chr2R 17715239 109 - 19596830 -------CGAUAUCGGGCCAAAACA---GAGAGAACAGCCUCUGU-----CAAAAGCCA-AGUGGCCAUUGGUAAUUGGAA-AA--GCCUGACAGCCAAUGGUGAUUAAAGUUAAUUGAACUAACACC -------((....))(((....(((---(((........))))))-----.....))).-(((.(((((((((..(..(..-..--..)..)..))))))))).)))...((((.......))))... ( -30.50, z-score = -2.50, R) >dp4.chr3 16626510 126 + 19779522 CACUUUGGGCUCACAAUCGAAAGGAUUCUGAAAAGAAGCCUUUCUGCCGAUGAAUGCUGCGGUAGAAGGGUACUCUUAGAAGGAUGGUCUGGCAUGGCUCAUGGGUCAAAGCUCACCAAACUUAUA-- ..((..(((.......((....)).((((....))))((((((((((((..........)))))))))))).)))..))(((..((((..(((.(((((....)))))..))).))))..)))...-- ( -40.30, z-score = -1.84, R) >consensus _______CGAUAUCGGGCCAAAACA___GAGAGAACAGCCUCUGU_____CAAAAGCCA_AGUGGCAAUUGGUAAUUGGAAAAA__GCCUGACAGCCAAUGGUGAUUAAAGUUAAUUGAACUAACACC .............................((((......)))).......(((..((...(((..(.((((((..((((.........))))..)))))).)..)))...))...))).......... ( -9.23 = -7.10 + -2.13)

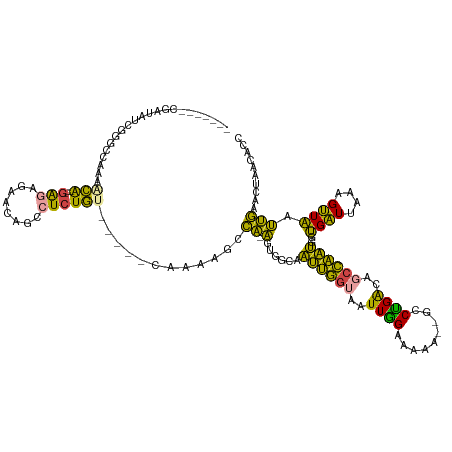
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:12 2011