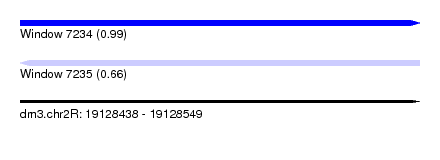
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,128,438 – 19,128,549 |
| Length | 111 |
| Max. P | 0.987583 |

| Location | 19,128,438 – 19,128,549 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.07 |
| Shannon entropy | 0.29783 |
| G+C content | 0.58774 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -32.96 |
| Energy contribution | -33.63 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.987583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19128438 111 + 21146708 CUCCAUCGCAGGGCGGGGAACUUGAACCGAGAUCCCUAUUGGGGUCAACUUCCCGGGGGCUCCGACACACGCAUCGCAUGGUAUCUCUGUACGAAGCUCCCAUUUAACACA ..........((((((..........))).((((((....)))))).....)))(((((((.(((((.....(((....))).....))).)).))))))).......... ( -33.90, z-score = -0.21, R) >droSim1.chr2R 17710491 92 + 19596830 ----------------CUCCUUCGCAGGGAGACCCUU-UUGGGGUCAACUUCCCGGGGGCUCCGACACACGCAUCGCAUGGUAUCUCUGUACGGAGCUCCCAUUUCCCG-- ----------------..........(((((((((..-...)))))........(((((((((((((.....(((....))).....))).))))))))))...)))).-- ( -39.30, z-score = -3.46, R) >droSec1.super_9 2422093 92 + 3197100 ----------------CUCCUUCGCAGGGAGACCCUU-UUGGGGUCAAAUUCCCGGGGGCUCCGACACACGCAUCGCAUGGUAUCUCUGUACGGAGCUCUCAUUUCCCG-- ----------------..........(((((((((..-...)))))........(((((((((((((.....(((....))).....))).))))))))))...)))).-- ( -36.30, z-score = -2.88, R) >consensus ________________CUCCUUCGCAGGGAGACCCUU_UUGGGGUCAACUUCCCGGGGGCUCCGACACACGCAUCGCAUGGUAUCUCUGUACGGAGCUCCCAUUUCCCG__ ..........................((((((((((....))))))........(((((((((((((.....(((....))).....))).))))))))))...))))... (-32.96 = -33.63 + 0.67)

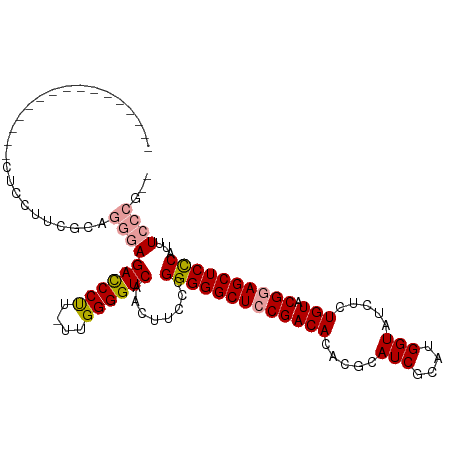

| Location | 19,128,438 – 19,128,549 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.07 |
| Shannon entropy | 0.29783 |
| G+C content | 0.58774 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -27.91 |
| Energy contribution | -28.58 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.655491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19128438 111 - 21146708 UGUGUUAAAUGGGAGCUUCGUACAGAGAUACCAUGCGAUGCGUGUGUCGGAGCCCCCGGGAAGUUGACCCCAAUAGGGAUCUCGGUUCAAGUUCCCCGCCCUGCGAUGGAG .(((......(((.((((((.(((.......(((((...)))))))))))))).)))(((((.((((((((....))(....))).)))).))))))))((......)).. ( -35.41, z-score = -0.38, R) >droSim1.chr2R 17710491 92 - 19596830 --CGGGAAAUGGGAGCUCCGUACAGAGAUACCAUGCGAUGCGUGUGUCGGAGCCCCCGGGAAGUUGACCCCAA-AAGGGUCUCCCUGCGAAGGAG---------------- --.((((..((((.((((((.(((.......(((((...)))))))))))))).)))).......(((((...-..)))))))))..........---------------- ( -36.41, z-score = -2.07, R) >droSec1.super_9 2422093 92 - 3197100 --CGGGAAAUGAGAGCUCCGUACAGAGAUACCAUGCGAUGCGUGUGUCGGAGCCCCCGGGAAUUUGACCCCAA-AAGGGUCUCCCUGCGAAGGAG---------------- --.(((......(.((((((.(((.......(((((...))))))))))))))))))((((....(((((...-..)))))))))..........---------------- ( -30.71, z-score = -1.23, R) >consensus __CGGGAAAUGGGAGCUCCGUACAGAGAUACCAUGCGAUGCGUGUGUCGGAGCCCCCGGGAAGUUGACCCCAA_AAGGGUCUCCCUGCGAAGGAG________________ ...((((..((((.((((((.(((.......(((((...)))))))))))))).)))).......(((((......))))))))).......................... (-27.91 = -28.58 + 0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:11 2011