| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,127,173 – 19,127,275 |
| Length | 102 |
| Max. P | 0.837281 |

| Location | 19,127,173 – 19,127,275 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 74.06 |
| Shannon entropy | 0.48383 |
| G+C content | 0.45783 |
| Mean single sequence MFE | -27.76 |
| Consensus MFE | -13.28 |
| Energy contribution | -15.17 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.534129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
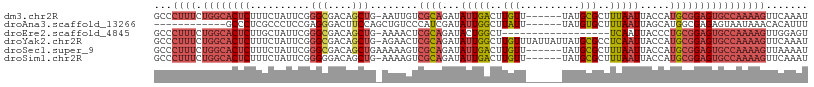
>dm3.chr2R 19127173 102 + 21146708 AUUUGAACUUUUGGCACUCCGCAUGGUAAUUAAAGCGCAUA------AACAAGUCAAUAUCUGCGACAAUU-CAGCUGUCGCCCGAAUAGAAAGAGUGCCAGAAAGGGC ........((((((((((((((............)))....------...............((((((...-....))))))...........)))))))))))..... ( -26.40, z-score = -1.28, R) >droAna3.scaffold_13266 8220329 91 - 19884421 AAAUGUGUUUAUUACUCUCGCCAUGCUAAUUAAAGCACAUA------AAUAAGCCAAUAUCGAUGGGACAGCUGGAAGUCCCUCGGAGGGCGAGGGC------------ ..............((((((((.((((......))))....------............((((.(((((........)))))))))..)))))))).------------ ( -30.50, z-score = -2.91, R) >droEre2.scaffold_4845 20495986 90 + 22589142 ACUCCAACUUUUGGCACUCCGCAGGGUAAUUGA------------------AGCCAGUAUCUGCGAGUUUU-CAGCUGUCGCCCGAAUAGCAAGAGUGCCAGAAAGGGC ...((...(((((((((((((((((...((((.------------------...)))).))))))......-..((((((....).)))))..)))))))))))..)). ( -33.40, z-score = -2.76, R) >droYak2.chr2R 19092267 108 + 21139217 AUUUGAACUUUUGGCACUCCGCAUGGUAAUUGAGGCGCAUAAUAAUAAACAAGCCAAUAUCUGCGAGUUCU-CAGCUGUCGCCCGAAUAGAAAGAGUGCCAGAAAGGGC ........(((((((((((((((.((((.....(((................)))..)))))))).((((.-..((....))..)))).....)))))))))))..... ( -28.59, z-score = -0.82, R) >droSec1.super_9 2420832 103 + 3197100 AUUUUAACUUUUGGCACUCCGCAUGGUAAUUAAAGCGCAUA------AACAAGUCAAUAUCUGCGACUUUUUCAGCUGUCGCCCGAAUAGAAAGAGUGCCAGAAAGGGC ........(((((((((((.(((((((....((((((((..------..............)))).))))....))))).)).(.....)...)))))))))))..... ( -25.39, z-score = -0.97, R) >droSim1.chr2R 17709241 102 + 19596830 AUUUGAACUUUUGGCACUCCGCAUGGUAAUUAAAGCGCAUA------AACAAGUCAAUAUCUGCGACUUUU-CAGCUGUCCCCCGAAUAGAAAGAGUGCCAGAAAGGGC ........(((((((((((((((.((((.............------..........)))))))).((.((-(...........))).))...)))))))))))..... ( -22.30, z-score = -0.09, R) >consensus AUUUGAACUUUUGGCACUCCGCAUGGUAAUUAAAGCGCAUA______AACAAGCCAAUAUCUGCGACUUUU_CAGCUGUCGCCCGAAUAGAAAGAGUGCCAGAAAGGGC ........(((((((((((((((.((((.............................)))))))).............((....)).......)))))))))))..... (-13.28 = -15.17 + 1.89)



| Location | 19,127,173 – 19,127,275 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 74.06 |
| Shannon entropy | 0.48383 |
| G+C content | 0.45783 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -14.63 |
| Energy contribution | -15.70 |
| Covariance contribution | 1.07 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.837281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19127173 102 - 21146708 GCCCUUUCUGGCACUCUUUCUAUUCGGGCGACAGCUG-AAUUGUCGCAGAUAUUGACUUGUU------UAUGCGCUUUAAUUACCAUGCGGAGUGCCAAAAGUUCAAAU (..((((.((((((((..((...((..(((((((...-..))))))).))....))......------....(((............)))))))))))))))..).... ( -29.70, z-score = -2.14, R) >droAna3.scaffold_13266 8220329 91 + 19884421 ------------GCCCUCGCCCUCCGAGGGACUUCCAGCUGUCCCAUCGAUAUUGGCUUAUU------UAUGUGCUUUAAUUAGCAUGGCGAGAGUAAUAAACACAUUU ------------((.((((((...((((((((........))))).))).............------...(((((......))))))))))).))............. ( -29.10, z-score = -3.31, R) >droEre2.scaffold_4845 20495986 90 - 22589142 GCCCUUUCUGGCACUCUUGCUAUUCGGGCGACAGCUG-AAAACUCGCAGAUACUGGCU------------------UCAAUUACCCUGCGGAGUGCCAAAAGUUGGAGU .((((((.((((((((......(((((.(....))))-))....(((((.........------------------.........)))))))))))))))))..))... ( -26.37, z-score = -0.51, R) >droYak2.chr2R 19092267 108 - 21139217 GCCCUUUCUGGCACUCUUUCUAUUCGGGCGACAGCUG-AGAACUCGCAGAUAUUGGCUUGUUUAUUAUUAUGCGCCUCAAUUACCAUGCGGAGUGCCAAAAGUUCAAAU (..((((.(((((((((....(((.(((((.((..((-(((((..((........))..)))).)))...))))))).)))........)))))))))))))..).... ( -29.80, z-score = -1.39, R) >droSec1.super_9 2420832 103 - 3197100 GCCCUUUCUGGCACUCUUUCUAUUCGGGCGACAGCUGAAAAAGUCGCAGAUAUUGACUUGUU------UAUGCGCUUUAAUUACCAUGCGGAGUGCCAAAAGUUAAAAU ...((((.(((((((((....(((..((((.((..((((.((((((.......)))))).))------))))))))..)))........)))))))))))))....... ( -28.10, z-score = -1.71, R) >droSim1.chr2R 17709241 102 - 19596830 GCCCUUUCUGGCACUCUUUCUAUUCGGGGGACAGCUG-AAAAGUCGCAGAUAUUGACUUGUU------UAUGCGCUUUAAUUACCAUGCGGAGUGCCAAAAGUUCAAAU (..((((.((((((((...((.(((((.......)))-)).)).((((...(((((..(((.------...)))..))))).....))))))))))))))))..).... ( -27.90, z-score = -1.26, R) >consensus GCCCUUUCUGGCACUCUUUCUAUUCGGGCGACAGCUG_AAAACUCGCAGAUAUUGACUUGUU______UAUGCGCUUUAAUUACCAUGCGGAGUGCCAAAAGUUCAAAU ........((((((((..........(((....)))........((((...(((((..(((..........)))..))))).....))))))))))))........... (-14.63 = -15.70 + 1.07)
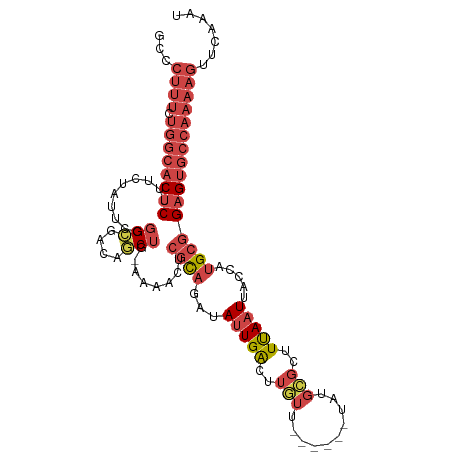

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:09 2011