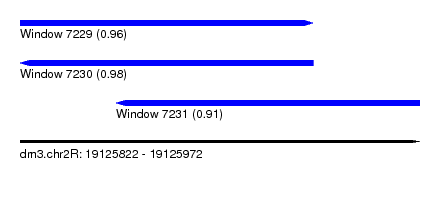
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,125,822 – 19,125,972 |
| Length | 150 |
| Max. P | 0.980004 |

| Location | 19,125,822 – 19,125,932 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 71.04 |
| Shannon entropy | 0.55352 |
| G+C content | 0.49669 |
| Mean single sequence MFE | -34.93 |
| Consensus MFE | -16.11 |
| Energy contribution | -16.58 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
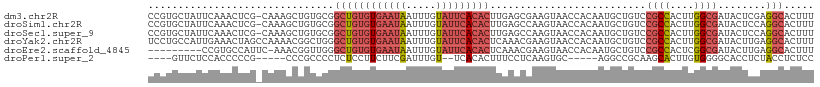
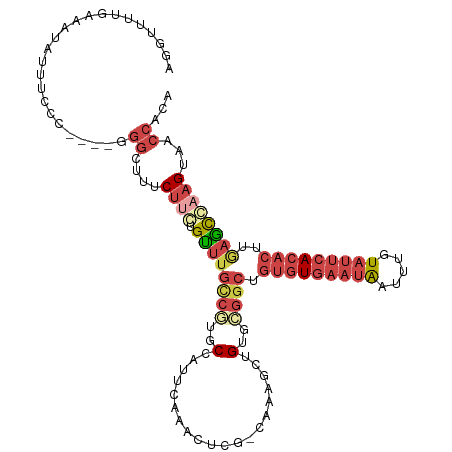
>dm3.chr2R 19125822 110 + 21146708 AAAGUGCCUCGAGUAUCGCCAAGUGGCGGACAGCAUUGUGGUUACUUCGCUCAAGUGUGAAUACAAAUUAUUCACACAGCCGCACAGCUUUG-CGAGUUUGAAUAGCACGG ...((((.((((...((((.((((.((((.(......((((.....))))....(((((((((.....))))))))).)))))...)))).)-)))..))))...)))).. ( -39.00, z-score = -3.02, R) >droSim1.chr2R 17707875 110 + 19596830 AAAGUGCCUGGAGUAUCGCCAAGUGGCGGACAGCAUUGUGGUUACUUGGCUCAAGUGUGAAUACAAAUUAUUCACACAGCCGCACAGCUUUG-CGAGUUUGAAUAGCACGG ...((((..........((((((((((.(.(......)).))))))))))(((((((((((((.....))))))))).((((((......))-)).))))))...)))).. ( -38.70, z-score = -2.42, R) >droSec1.super_9 2419458 110 + 3197100 AAAGUGCCUGGAGUAUCGCCAAGUGGCGGACAGCAUUGUGGUUACUUGGCUCAAGUGUGAAUACAAAUUAUUCACACAGCCGCACAGCUUUG-CGAGUUUGAAUAGCACGG ...((((..........((((((((((.(.(......)).))))))))))(((((((((((((.....))))))))).((((((......))-)).))))))...)))).. ( -38.70, z-score = -2.42, R) >droYak2.chr2R 19090864 111 + 21139217 AAAGUGCCUCAAGUAUCGCCAAGUGGCGGACAGCAUUGUGGUUACUUCGUUUGAGUGUGAAUACAAAUUAUUCACACAGCCCAGCCGUUUUGGCUAGUUUCAAUGGCAGGA .(((((((.(((((.(((((....)))))...)).))).)).))))).......(((((((((.....)))))))))...((.((((((..((....))..)))))).)). ( -34.20, z-score = -1.66, R) >droEre2.scaffold_4845 20494633 101 + 22589142 AAAGUGCCUCAAGUAUCGCCGAGUGGCGGACAGCAUUGUGGUUACUUCGUUUGAGUGUGAAUACAAAUUAUUCACACAGCCCAACCGUUU-GAAUGGCACGG--------- ...(((((((((((.(((((....)))))))....(((.((((...((....))(((((((((.....))))))))))))))))....))-))..)))))..--------- ( -32.90, z-score = -2.27, R) >droPer1.super_2 5680406 95 - 9036312 GGAGAGGUAGAGGUGCCCCACAAGUGCUUGCGGCCU-----GCACUUGAGGAAAGUGUGA--ACAAAUCGAAGAAGGAGAGGGGCGGG-----CGGGGGUGGAGAAC---- .....((((....))))((((...(((((((..(((-----((((((.....))))))..--.....((......))..))).)))))-----))...)))).....---- ( -26.10, z-score = -0.42, R) >consensus AAAGUGCCUCAAGUAUCGCCAAGUGGCGGACAGCAUUGUGGUUACUUCGCUCAAGUGUGAAUACAAAUUAUUCACACAGCCGCACAGCUUUG_CGAGUUUGAAUAGCACGG ..........(((((..((((....((.....))....))))))))).......(((((((((.....))))))))).................................. (-16.11 = -16.58 + 0.48)
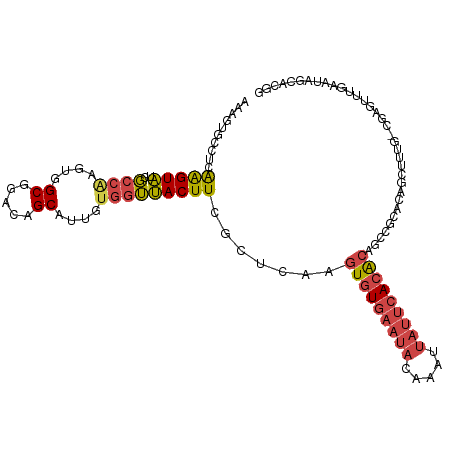
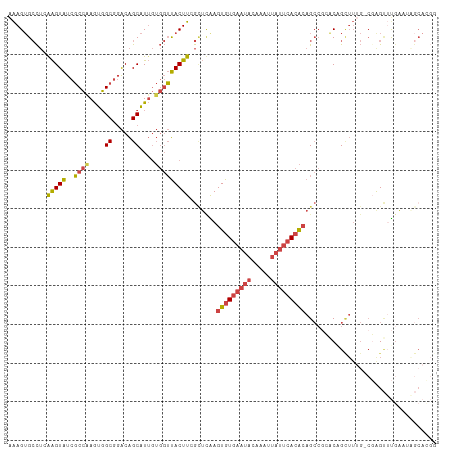
| Location | 19,125,822 – 19,125,932 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.04 |
| Shannon entropy | 0.55352 |
| G+C content | 0.49669 |
| Mean single sequence MFE | -31.99 |
| Consensus MFE | -13.07 |
| Energy contribution | -14.35 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
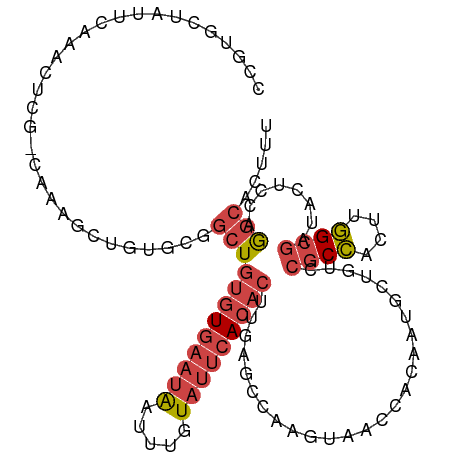
>dm3.chr2R 19125822 110 - 21146708 CCGUGCUAUUCAAACUCG-CAAAGCUGUGCGGCUGUGUGAAUAAUUUGUAUUCACACUUGAGCGAAGUAACCACAAUGCUGUCCGCCACUUGGCGAUACUCGAGGCACUUU ..(((((........(((-(.(((.((.(((((.(((((((((.....)))))))))...((((..((....))..))))).))))))))).)))).......)))))... ( -34.46, z-score = -2.08, R) >droSim1.chr2R 17707875 110 - 19596830 CCGUGCUAUUCAAACUCG-CAAAGCUGUGCGGCUGUGUGAAUAAUUUGUAUUCACACUUGAGCCAAGUAACCACAAUGCUGUCCGCCACUUGGCGAUACUCCAGGCACUUU ..(((((..((((..(((-((......)))))..(((((((((.....)))))))))))))(((((((....(((....))).....))))))).........)))))... ( -34.30, z-score = -2.40, R) >droSec1.super_9 2419458 110 - 3197100 CCGUGCUAUUCAAACUCG-CAAAGCUGUGCGGCUGUGUGAAUAAUUUGUAUUCACACUUGAGCCAAGUAACCACAAUGCUGUCCGCCACUUGGCGAUACUCCAGGCACUUU ..(((((..((((..(((-((......)))))..(((((((((.....)))))))))))))(((((((....(((....))).....))))))).........)))))... ( -34.30, z-score = -2.40, R) >droYak2.chr2R 19090864 111 - 21139217 UCCUGCCAUUGAAACUAGCCAAAACGGCUGGGCUGUGUGAAUAAUUUGUAUUCACACUCAAACGAAGUAACCACAAUGCUGUCCGCCACUUGGCGAUACUUGAGGCACUUU ...((((.((((..((((((.....))))))...(((((((((.....)))))))))))))...(((((...(((....))).((((....)))).)))))..)))).... ( -36.70, z-score = -3.84, R) >droEre2.scaffold_4845 20494633 101 - 22589142 ---------CCGUGCCAUUC-AAACGGUUGGGCUGUGUGAAUAAUUUGUAUUCACACUCAAACGAAGUAACCACAAUGCUGUCCGCCACUCGGCGAUACUUGAGGCACUUU ---------..(((((..((-((((((((((((((((((((((.....)))))))))((....)))))..)))....))))).((((....))))....)))))))))... ( -31.80, z-score = -2.53, R) >droPer1.super_2 5680406 95 + 9036312 ----GUUCUCCACCCCCG-----CCCGCCCCUCUCCUUCUUCGAUUUGU--UCACACUUUCCUCAAGUGC-----AGGCCGCAAGCACUUGUGGGGCACCUCUACCUCUCC ----..............-----...(((((...........((.....--))..........(((((((-----.........))))))).))))).............. ( -20.40, z-score = -1.60, R) >consensus CCGUGCUAUUCAAACUCG_CAAAGCUGUGCGGCUGUGUGAAUAAUUUGUAUUCACACUUGAGCCAAGUAACCACAAUGCUGUCCGCCACUUGGCGAUACUCCAGGCACUUU ...............................((((((((((((.....)))))))))..........................((((....))))........)))..... (-13.07 = -14.35 + 1.28)
| Location | 19,125,858 – 19,125,972 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 66.51 |
| Shannon entropy | 0.63625 |
| G+C content | 0.43339 |
| Mean single sequence MFE | -27.39 |
| Consensus MFE | -11.40 |
| Energy contribution | -13.57 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.910656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
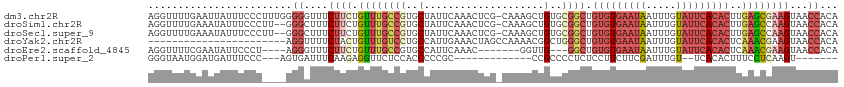
>dm3.chr2R 19125858 114 - 21146708 AGGUUUUGAAUUAUUUCCCUUUGGGGGUUUCUUCUGUUUGCCGUGCUAUUCAAACUCG-CAAAGCUGUGCGGCUGUGUGAAUAAUUUGUAUUCACACUUGAGCGAAGUAACCACA ................(((...)))((((.((((.((((((((..(............-.......)..)))).(((((((((.....)))))))))..)))))))).))))... ( -33.71, z-score = -2.21, R) >droSim1.chr2R 17707911 112 - 19596830 AGGUUUUGAAAUAUUUCCCUU--GGGCUUUCUUCUGUUUGCCGUGCUAUUCAAACUCG-CAAAGCUGUGCGGCUGUGUGAAUAAUUUGUAUUCACACUUGAGCCAAGUAACCACA .((((..(((....))).(((--((.(((..........((((..(............-.......)..)))).(((((((((.....)))))))))..)))))))).))))... ( -30.01, z-score = -1.68, R) >droSec1.super_9 2419494 112 - 3197100 AGGUUUUGAAAUAUUUCCCUU--GGGCUUUCUUCUGUUUGCCGUGCUAUUCAAACUCG-CAAAGCUGUGCGGCUGUGUGAAUAAUUUGUAUUCACACUUGAGCCAAGUAACCACA .((((..(((....))).(((--((.(((..........((((..(............-.......)..)))).(((((((((.....)))))))))..)))))))).))))... ( -30.01, z-score = -1.68, R) >droYak2.chr2R 19090900 92 - 21139217 -----------------------AGGUUUUCUACUGUUUGUCCUGCCAUUGAAACUAGCCAAAACGGCUGGGCUGUGUGAAUAAUUUGUAUUCACACUCAAACGAAGUAACCACA -----------------------.((((..((..((((((..............((((((.....))))))...(((((((((.....))))))))).)))))).)).))))... ( -28.10, z-score = -3.07, R) >droEre2.scaffold_4845 20494669 101 - 22589142 AGGUUUUCGAAUAUUCCCU----AGGGUUUCUUCUGUUUGCCGUGCCAUUCAAAC-------GGUUG---GGCUGUGUGAAUAAUUUGUAUUCACACUCAAACGAAGUAACCACA ...................----..((((.((((.(((((((..(((........-------)))..---))).(((((((((.....)))))))))..)))))))).))))... ( -28.80, z-score = -2.32, R) >droPer1.super_2 5680444 90 + 9036312 GGGUAAUGGAUGAUUUCCC---AGUGAUUUCAAGAGGUUCUCCACCCCCGC-------------CCGCCCCUCUCCUUCUUCGAUUUGU--UCACACUUUCCUCAAGU------- ((((..((((.((((((..---..((....)).)))))).)))).....))-------------)).......................--.................------- ( -13.70, z-score = 1.33, R) >consensus AGGUUUUGAAAUAUUUCCC____GGGCUUUCUUCUGUUUGCCGUGCCAUUCAAACUCG_CAAAGCUGUGCGGCUGUGUGAAUAAUUUGUAUUCACACUUGAGCCAAGUAACCACA ........................((....((((.((((((((..(....................)..)))).(((((((((.....)))))))))..))))))))...))... (-11.40 = -13.57 + 2.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:08 2011