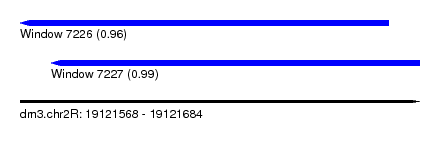
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,121,568 – 19,121,684 |
| Length | 116 |
| Max. P | 0.985884 |

| Location | 19,121,568 – 19,121,675 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 91.43 |
| Shannon entropy | 0.14437 |
| G+C content | 0.40505 |
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -23.58 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
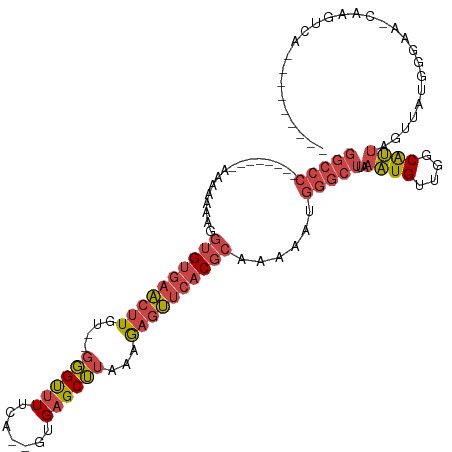
>dm3.chr2R 19121568 107 - 21146708 AAAAAAAAAAAGGUGUGAACUUGU--GGGUUUUCAGUGAGCUUAAAGAGUUCACGCAAAAAUGGGCUUAAAUGAUGGCAUUAGUUAUGGGAACCAAAUCAUCGGAUGAA ............((((((((((.(--((((((.....)))))))..)))))))))).....(((..((..(((((.......)))))..)).))).............. ( -23.70, z-score = -1.60, R) >droEre2.scaffold_4845 20490436 98 - 22589142 --------AAAGGUGUGAACUUGU--GGGCAUUCAGUGAGCUUAAAGAGUUCACGCAAAAAUGGGCUUAAAUGUUGGCAUUAGUUAUGGGAA-CAAGCCAUCGGCUGAA --------....((((((((((.(--((((.........)))))..))))))))))........(((........))).((((((((((...-....)))).)))))). ( -27.30, z-score = -1.56, R) >droYak2.chr2R 19086453 101 - 21139217 -------AAAAGGUGUGAACUUGUGGGGGUUUUCAGUGAGCUUUAAGAGUUCACGCAAAAAUGGGCUUAAAUGUUGGCAUUAGUUAUGGGAA-CAAGCCAUCGGAUGAA -------.....((((((((((...(((((((.....)))))))..)))))))))).......(((((.((((....))))......(....-)))))).......... ( -28.50, z-score = -2.44, R) >droSec1.super_9 2415176 98 - 3197100 --------AAAGGUGUGAACUUGU--GGGUUUUCAGUGAGCUUAAAGAGUUCACGCAAAAAUGGGCUUAAAUGGUGGCAUUAGUUAUGGGAA-CAAGUCAUCGGAUGAA --------....((((((((((.(--((((((.....)))))))..)))))))))).......(((((...(.(((((....))))).)...-.))))).......... ( -26.20, z-score = -2.55, R) >droSim1.chr2R 17703606 98 - 19596830 --------AAAAGUGUGAACUUGU--GGGUUUUCAGUGAGCUUAAAGAGUUCACGCAAAAAUGGGCUUAAAUGGUGGCAUUAGUUAUGGGGA-CAAGUCAUCGGAUGAA --------....((((((((((.(--((((((.....)))))))..)))))))))).......(((((.....(((((....)))))(....-)))))).......... ( -27.00, z-score = -2.85, R) >consensus ________AAAGGUGUGAACUUGU__GGGUUUUCAGUGAGCUUAAAGAGUUCACGCAAAAAUGGGCUUAAAUGGUGGCAUUAGUUAUGGGAA_CAAGUCAUCGGAUGAA ............((((((((((....((((((.....))))))...)))))))))).......(((((...(.(((((....))))).).....))))).......... (-23.58 = -23.90 + 0.32)


| Location | 19,121,577 – 19,121,684 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.48 |
| Shannon entropy | 0.47756 |
| G+C content | 0.41476 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -16.59 |
| Energy contribution | -17.35 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.985884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19121577 107 - 21146708 GGCCCAAAAAAAAAAAAAAAGGUGUGAACUUGU--GGGUUUUCA--GUGAGCUUAAAGAGUUCACGCAAAAAUGGGCUUAAAUGAUGGCAUUAGUUAUGGGAACCAAAUCA--------- ((((((...............((((((((((.(--((((((...--..)))))))..))))))))))........(((........)))........))))..))......--------- ( -28.10, z-score = -2.36, R) >droEre2.scaffold_4845 20490445 98 - 22589142 GGCCC--------AAAAAAAGGUGUGAACUUGU--GGGCAUUCA--GUGAGCUUAAAGAGUUCACGCAAAAAUGGGCUUAAAUGUUGGCAUUAGUUAUGGGAA-CAAGCCA--------- (((((--------(.......((((((((((.(--((((.....--....)))))..)))))))))).....)))))).......((((.((.(((.....))-)))))))--------- ( -30.60, z-score = -2.32, R) >droYak2.chr2R 19086462 101 - 21139217 GGCCC-------AAAAAAAAGGUGUGAACUUGUGGGGGUUUUCA--GUGAGCUUUAAGAGUUCACGCAAAAAUGGGCUUAAAUGUUGGCAUUAGUUAUGGGAA-CAAGCCA--------- (((((-------(........((((((((((...(((((((...--..)))))))..)))))))))).....)))))).......((((.((.(((.....))-)))))))--------- ( -32.32, z-score = -2.65, R) >droSec1.super_9 2415185 98 - 3197100 GGCCC--------AAAAAAAGGUGUGAACUUGU--GGGUUUUCA--GUGAGCUUAAAGAGUUCACGCAAAAAUGGGCUUAAAUGGUGGCAUUAGUUAUGGGAA-CAAGUCA--------- (((((--------(.......((((((((((.(--((((((...--..)))))))..)))))))))).....))))))....(.(((((....))))).)...-.......--------- ( -29.90, z-score = -2.80, R) >droSim1.chr2R 17703615 98 - 19596830 GGCCC--------AAGAAAAAGUGUGAACUUGU--GGGUUUUCA--GUGAGCUUAAAGAGUUCACGCAAAAAUGGGCUUAAAUGGUGGCAUUAGUUAUGGGGA-CAAGUCA--------- (((((--------(.......((((((((((.(--((((((...--..)))))))..)))))))))).....))))))......(((((....)))))(....-)......--------- ( -29.90, z-score = -2.69, R) >droVir3.scaffold_12875 3105430 104 - 20611582 ------------ACCUUAAAUCAGAGCUCUUGUCGUUGUUUCCAUUGUGAGCACGCGACAACAACAUGAAACU----UAAGGUGUGAGCGCAAUUCAAGGUAAUAAAUUUGAAGAAGCCC ------------(((((((.(((......((((((((((((.......))))).))))))).....)))...)----))))))....((....((((((........))))))...)).. ( -23.20, z-score = -0.68, R) >consensus GGCCC________AAAAAAAGGUGUGAACUUGU__GGGUUUUCA__GUGAGCUUAAAGAGUUCACGCAAAAAUGGGCUUAAAUGUUGGCAUUAGUUAUGGGAA_CAAGUCA_________ (((((................((((((((((....((((((.......))))))...))))))))))......)))))..((((....))))............................ (-16.59 = -17.35 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:45:05 2011