
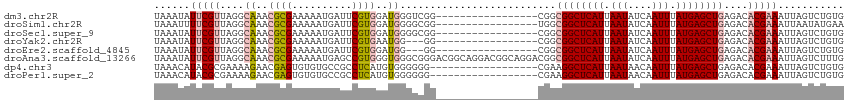
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,050,022 – 19,050,154 |
| Length | 132 |
| Max. P | 0.839182 |

| Location | 19,050,022 – 19,050,120 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.99 |
| Shannon entropy | 0.35479 |
| G+C content | 0.42992 |
| Mean single sequence MFE | -27.06 |
| Consensus MFE | -14.66 |
| Energy contribution | -15.16 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.730211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
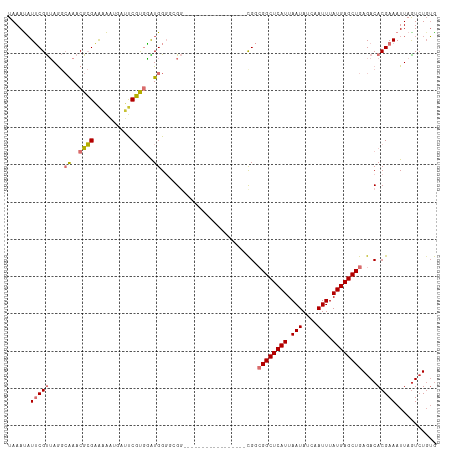
>dm3.chr2R 19050022 98 + 21146708 UAAAUAUUCGUUAGGCAAACGCGAAAAAUGAUUCGUGGAUGGGUCGG-----------------CGGCGGCUCAUUAAUAUCAAUUUAUGAGCUGAGACACGAAAUUAGUCUGUG ......(((((....((..((((((......))))))..)).(((..-----------------...((((((((.(((....))).)))))))).))))))))........... ( -26.00, z-score = -1.77, R) >droSim1.chr2R 17632388 98 + 19596830 UAAAUUUUCGUUAGGCAAACGCGAAAAAUGAUUCGUGGAUGGGGCGG-----------------UGGCGGCUCAUUAAUAUCAAUUUAUGAGCUGAGACACGAAAUUAAUAUGAA ..((((((((((...((..((((((......))))))..)).))))(-----------------((.((((((((.(((....))).))))))))...)))))))))........ ( -27.10, z-score = -3.10, R) >droSec1.super_9 2344438 98 + 3197100 UAAAUAUUCGUUAGGCAAACGCGAAAAAUGAUUCGUGGAUGGGGCGG-----------------CGGCGGCUCAUUAAUAUCAAUUUAUGAGCUGAGACACGAAAUUAGUCUGUG .......(((((...((..((((((......))))))..)).)))))-----------------...((((((((.(((....))).))))))))((((.........))))... ( -24.70, z-score = -1.31, R) >droYak2.chr2R 19013628 95 + 21139217 UAAAUAUUCGUUAGGCAAACGCGAAAAAUGAUUCGUGAAUGG---GG-----------------CGGCGGCUCAUUAAUAUCAAUUUAUGAGCUGAGACACGAAAUUAGUCUGUG .......(((((...((..((((((......))))))..)).---))-----------------)))((((((((.(((....))).))))))))((((.........))))... ( -22.70, z-score = -1.34, R) >droEre2.scaffold_4845 20416750 95 + 22589142 UAAAUAUUCGUUAGGCAAACGCGAAAAAUGAUUCGUGGAUGG---GG-----------------CGGCGGCUCAUUAAUAUCAAUUUAUGAGCUGAGACACGAAAUUAGUCUGUG .......(((((...((..((((((......))))))..)).---))-----------------)))((((((((.(((....))).))))))))((((.........))))... ( -24.70, z-score = -1.95, R) >droAna3.scaffold_13266 8147208 115 - 19884421 UAAAUAUUCGUUAGGCAAACGCGAAAAAUGAGCCGUGGGUGGGCGGGACGGCAGGACGGCAGGACGGCGGCUCAUUAAUAUCAAUUUAUGAGCUGAGACACGAAAUUAGUCUUUG ......(((((..(((..........((((((((((.(....((.(..(....)..).))....).))))))))))....(((.....)))))).....)))))........... ( -32.70, z-score = -1.93, R) >dp4.chr3 16524947 97 - 19779522 UAAACAUACGCGAAAAGAACGAGUGUGUGCCGCCUCAUGUGGGGGG------------------CGAAGGCUCAUUAAUAACAAUUUAUGAGCUGAGACACGAAAUUAGUCUGUG ...((((((.((.......)).))))))(((.((((....))))))------------------)...(((((((.(((....))).))))))).((((.........))))... ( -29.30, z-score = -2.64, R) >droPer1.super_2 5589922 97 - 9036312 UAAACAUACGCGAAAAGAACGAGUGUGUGCCGCCUCAUGUGGGGGG------------------CGAAGGCUCAUUAAUAACAAUUUAUGAGCUGAGACACGAAAUUAGUCUGUG ...((((((.((.......)).))))))(((.((((....))))))------------------)...(((((((.(((....))).))))))).((((.........))))... ( -29.30, z-score = -2.64, R) >consensus UAAAUAUUCGUUAGGCAAACGCGAAAAAUGAUUCGUGGAUGGGGCGG_________________CGGCGGCUCAUUAAUAUCAAUUUAUGAGCUGAGACACGAAAUUAGUCUGUG ......(((((....((..((((..........))))..))..........................((((((((.(((....))).))))))))....)))))........... (-14.66 = -15.16 + 0.50)

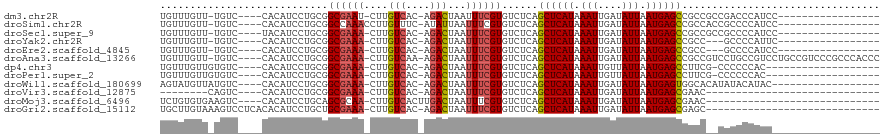
| Location | 19,050,058 – 19,050,154 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.65 |
| Shannon entropy | 0.38127 |
| G+C content | 0.44761 |
| Mean single sequence MFE | -20.53 |
| Consensus MFE | -14.39 |
| Energy contribution | -14.57 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.839182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
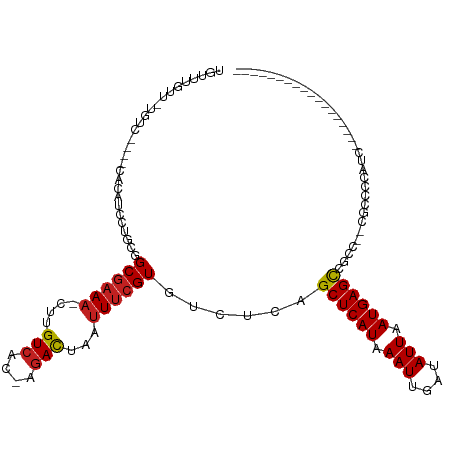
>dm3.chr2R 19050058 96 - 21146708 UGUUUGUU-UGUC----CACAUCCUGCGGCGAAU-CUUGUCAC-AGACUAAUUUCGUGUCUCAGCUCAUAAAUUGAUAUUAAUGAGCCGCCGCCGACCCAUCC----------------- .(..(((.-....----.)))..).((((((...-........-((((.........))))..((((((.(((....))).))))))))))))..........----------------- ( -22.70, z-score = -1.91, R) >droSim1.chr2R 17632424 97 - 19596830 UGUUUGUU-UGUC----CACAUCCUGCGGCCAAACCUUGUUUC-AUAUUAAUUUCGUGUCUCAGCUCAUAAAUUGAUAUUAAUGAGCCGCCACCGCCCCAUCC----------------- .(..(((.-....----.)))..).((((......((..((..-((((((((((.(((........))))))))))))).))..))......)))).......----------------- ( -16.10, z-score = -1.26, R) >droSec1.super_9 2344474 96 - 3197100 UGUUUGUU-UGUC----UACAUCCUGCGGCGAAA-CUUGUCAC-AGACUAAUUUCGUGUCUCAGCUCAUAAAUUGAUAUUAAUGAGCCGCCGCCGCCCCAUCC----------------- .(..(((.-....----.)))..).((((((...-........-((((.........))))..((((((.(((....))).))))))))))))..........----------------- ( -23.40, z-score = -2.31, R) >droYak2.chr2R 19013664 93 - 21139217 UGUUUGUU-UGUC----CACAUCCUGCGGCGAAA-CUUGUCAC-AGACUAAUUUCGUGUCUCAGCUCAUAAAUUGAUAUUAAUGAGCCGCC---GCCCCAUUC----------------- .(..(((.-....----.)))..).((((((...-........-((((.........))))..((((((.(((....))).))))))))))---)).......----------------- ( -22.70, z-score = -2.39, R) >droEre2.scaffold_4845 20416786 93 - 22589142 UGUUUGUU-UGUC----CACAUCCUGCGGCGAAA-CUUGUCAC-AGACUAAUUUCGUGUCUCAGCUCAUAAAUUGAUAUUAAUGAGCCGCC---GCCCCAUCC----------------- .(..(((.-....----.)))..).((((((...-........-((((.........))))..((((((.(((....))).))))))))))---)).......----------------- ( -22.70, z-score = -2.50, R) >droAna3.scaffold_13266 8147244 113 + 19884421 UGUUUGUU-UGUC----CACAUCCUGCGGCGAAA-CUUGUCAA-AGACUAAUUUCGUGUCUCAGCUCAUAAAUUGAUAUUAAUGAGCCGCCGUCCUGCCGUCCUGCCGUCCCGCCCACCC .(..(((.-....----.)))..).((((((...-........-((((.........))))..((((((.(((....))).))))))))))))...((.(..(....)..).))...... ( -21.40, z-score = -0.67, R) >dp4.chr3 16524985 94 + 19779522 UGUUUGUUGUGUC----CACAUCCUGCGGCGAAA-CUUGUCAC-AGACUAAUUUCGUGUCUCAGCUCAUAAAUUGUUAUUAAUGAGCCUUCG-CCCCCCAC------------------- .((..(.(((...----.))).)..))((((((.-........-((((.........))))..((((((.(((....))).)))))).))))-))......------------------- ( -21.00, z-score = -2.04, R) >droPer1.super_2 5589960 94 + 9036312 UGUUUGUUGUGUC----CACAUCCUGCGGCGAAA-CUUGUCAC-AGACUAAUUUCGUGUCUCAGCUCAUAAAUUGUUAUUAAUGAGCCUUCG-CCCCCCAC------------------- .((..(.(((...----.))).)..))((((((.-........-((((.........))))..((((((.(((....))).)))))).))))-))......------------------- ( -21.00, z-score = -2.04, R) >droWil1.scaffold_180699 2215211 96 + 2593675 AGUAUGUUAUGUC----CACAUCCUGCGGCGAAA-CUUGUCAC-AGACUAAUUUCGUGUCUCAGCUCAUAAAUUGAUAUUAAUGAGUGGCACAUAUACAUAC------------------ .((((((((((((----(((...(((.(((((((-...(((..-.)))...))))))..).))).((((.(((....))).)))))))).))))).))))))------------------ ( -25.50, z-score = -2.38, R) >droVir3.scaffold_12875 3025806 77 - 20611582 --------CAGUC----CACAUCCUGCGGCGAAA-CUUGUCAC-AGACUAAUUUCGUGUCUCAGCUCAUAAAUUGAUAUUAAUGAGCGAAC----------------------------- --------.....----...........((((((-...(((..-.)))...))))))......((((((.(((....))).))))))....----------------------------- ( -15.10, z-score = -0.64, R) >droMoj3.scaffold_6496 4701215 86 + 26866924 UCUGUGUGAAGUC----CACAUCCUGCAGCGCAA-CUUGUCACUUGACUAAUUUCGUGUCUCAGCUCAUAAAUUGAUAUUAAUGAGCGAAC----------------------------- ...(((((.....----)))))..((..((((..-...(((....))).......))))..))((((((.(((....))).))))))....----------------------------- ( -17.20, z-score = -0.26, R) >droGri2.scaffold_15112 2039608 89 + 5172618 UGCUUGUAAAGUCCUCACACAUCCUGCUGCGAAA-CUUGUCAC-AGACUAAUUUCGUGUCUCAGCUCAUAAAUUGAUAUUAAUGAGCGAGC----------------------------- .((((.......................((((((-...(((..-.)))...))))))......((((((.(((....))).))))))))))----------------------------- ( -17.60, z-score = -0.36, R) >consensus UGUUUGUU_UGUC____CACAUCCUGCGGCGAAA_CUUGUCAC_AGACUAAUUUCGUGUCUCAGCUCAUAAAUUGAUAUUAAUGAGCCGCC__CGCCCCAUC__________________ ............................((((((....(((....)))...))))))......((((((.(((....))).))))))................................. (-14.39 = -14.57 + 0.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:59 2011