| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,037,573 – 19,037,723 |
| Length | 150 |
| Max. P | 0.970788 |

| Location | 19,037,573 – 19,037,723 |
|---|---|
| Length | 150 |
| Sequences | 9 |
| Columns | 174 |
| Reading direction | reverse |
| Mean pairwise identity | 69.32 |
| Shannon entropy | 0.59819 |
| G+C content | 0.46700 |
| Mean single sequence MFE | -45.73 |
| Consensus MFE | -22.61 |
| Energy contribution | -23.04 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
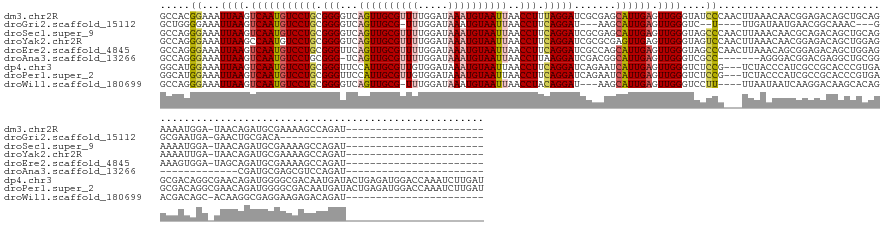
>dm3.chr2R 19037573 150 - 21146708 GCCACGGAAAUUAAGUCAAUGUCCUGCGGGGUCAGUUGCGUUUUGGAUAAAUGUAAUUAACCUUUAGGAUCGCGAGCAUUGAGUUGGGUAUCCCAACUUAAACAACGGAGACAGCUGCAGAAAAUGGA-UAACAGAUGCGAAAAGCCAGAU----------------------- .....((.......(((...((((((.(((((.((((((((((.....)))))))))).))))))))))).((.(((.((((((((((...)))))))))).....(....).)))))..........-.....)))........))....----------------------- ( -44.06, z-score = -2.21, R) >droGri2.scaffold_15112 2025860 126 + 5172618 GCUGGGGAAAUUAAGUCAAUGUCCUGCGGGGUCAGUUGCG-UUUGGAUAAAUGUAAUUAACCUUCAGGAU---AAGCAUUGAGUUGGGUC--U----UUGAUAAUGAACGGCAAAC---GGCGAAUGA-GAACUGCGACA---------------------------------- ((((......((((.(((((((((((.(((((.(((((((-(((....)))))))))).)))))))))..---..))))))).))))(((--(----((......))).)))...)---)))......-...........---------------------------------- ( -34.90, z-score = -1.32, R) >droSec1.super_9 2332016 150 - 3197100 GCCAGGGAAAUUAAGUCAAUGUCCUGCGGGGUCAGUUGCGUUUUGGAUAAAUGUAAUUAACCUUCAGGAUCGCGAGCAUUGAGUUGGGUAGCCCAACUUAAACAACGCAGACAGCUGCAGAAAAUGGA-UAACAGAUGCGAAAAGCCAGAU----------------------- ....((........(((...((((((.(((((.((((((((((.....)))))))))).))))))))))).(((....((((((((((...))))))))))....))).)))...((((.....((..-...))..)))).....))....----------------------- ( -46.80, z-score = -2.51, R) >droYak2.chr2R 19001139 150 - 21139217 GCCAGGGAAAUUAAGCCAAUGUCCUGCGGGGUCAGUUGCGUUUUGGAUAAAUGUAAUUAACCUUCAGGAUCGCGCGAGUUGAGUUGGGUAGUCCAACUUAAACAACGGAGACAGCUGGAGAAAAUUGA-UAACAGAUGCGAAAAGCCAGAU----------------------- .((((.........((....((((((.(((((.((((((((((.....)))))))))).)))))))))))...))...(((((((((.....))))))))).....(....)..))))..........-........((.....)).....----------------------- ( -41.30, z-score = -1.07, R) >droEre2.scaffold_4845 20404706 150 - 22589142 GCCAGGGAAAUUAAGUCAAUGUCCUGCGGGUUCAGUUGCGUUUUGGAUAAAUGUAAUUAACCUUCAGGAUCGCCAGCAUUGAGUUGGGUAGCCCAACUUAAACAGCGGAGACAGCUGGAGAAAGUGGA-UAGCAGAUGCGAAAAGCCAGAU----------------------- ....((........(((....(((((.(((((.((((((((((.....))))))))))))))).)))))((.(((((.((((((((((...)))))))))).....(....).))))).)).....))-).((....))......))....----------------------- ( -51.40, z-score = -3.25, R) >droAna3.scaffold_13266 8135555 130 + 19884421 GCCAGGGAAAUUAAGUCAAUGUCCUGCGGG-UCAGUUGCGUUUUGGAUAAAUGUAAUUAACCUUAAGGAUCGACGGCAUUGAGUUGGGUCGCC-------AGGGACGGACGAGGCUGCGG-------------CGAUGCGAGCGUCCAGAU----------------------- .((..((...((((.((((((((((..(((-(.((((((((((.....)))))))))).))))..))).......))))))).))))....))-------..))..(((((....((((.-------------...))))..)))))....----------------------- ( -42.21, z-score = -0.60, R) >dp4.chr3 16514984 171 + 19779522 GGCAUGGAAAUUAAGUCAAUGUCCUGCGGGUUCCAUUGCGUUGUGGAUAAAUGUAAUUAACCUUCAGGAUCAGAAUCAUUGAGUUGGGUCUCCG---UCUACCCAUCGCCGCACCCGUGAGCGACAGGCGAACAGAUGGGGCGACAAUGAUACUGAGAUGGACCAAAUCUUGAU ....(((.......(((....(((((.(((((..((((((((.......)))))))).))))).)))))((((.(((((((...(((((.....---...)))))(((((.((.(.((..((.....))..)).).)).)))))))))))).)))))))...)))......... ( -55.70, z-score = -1.18, R) >droPer1.super_2 5578859 171 + 9036312 GGCAUGGAAAUUAAGUCAAUGUCCUGCGGGUUCCAUUGCGUUGUGGAUAAAUGUAAUUAACCUUCAGGAUCAGAAUCAUUGAGUUGGGUCUCCG---UCUACCCAUCGCCGCACCCGUGAGCGACAGGCGAACAGAUGGGGCGACAAUGAUACUGAGAUGGACCAAAUCUUGAU ....(((.......(((....(((((.(((((..((((((((.......)))))))).))))).)))))((((.(((((((...(((((.....---...)))))(((((.((.(.((..((.....))..)).).)).)))))))))))).)))))))...)))......... ( -55.70, z-score = -1.18, R) >droWil1.scaffold_180699 2200191 142 + 2593675 GCCAGGGAAAUUAAGUCAAUGUCCUGCGGGGUCAGUUGCG-UUUGGAUAAAUGUAAUUAACCUACAGGAU---AAGCAUUGAGUUGGGUCCUU----UUAAUAAUCAAGGACAAGCACAGACGACAGC-ACAAGGCGAGGAAGAGACAGAU----------------------- (((..(........(((..(((((((..((((.(((((((-(((....)))))))))).)))).))))))---).....((.(((..((((((----.........)))))).))).)))))......-.)..)))...............----------------------- ( -39.46, z-score = -2.42, R) >consensus GCCAGGGAAAUUAAGUCAAUGUCCUGCGGGGUCAGUUGCGUUUUGGAUAAAUGUAAUUAACCUUCAGGAUCGCAAGCAUUGAGUUGGGUCGCCC___UUAAACAACGGAGACAGCUGCAGAAAAUAGA_UAACAGAUGCGAAAAGCCAGAU_______________________ .....((...((((.(((((((((((.(((...((((((((((.....))))))))))..))).))))).......)))))).))))....))................................................................................. (-22.61 = -23.04 + 0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:56 2011