
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,028,681 – 19,028,772 |
| Length | 91 |
| Max. P | 0.778380 |

| Location | 19,028,681 – 19,028,772 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 74.54 |
| Shannon entropy | 0.48932 |
| G+C content | 0.46502 |
| Mean single sequence MFE | -25.69 |
| Consensus MFE | -12.50 |
| Energy contribution | -12.59 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.778380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
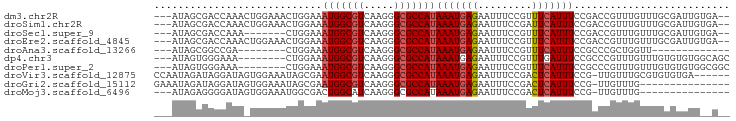
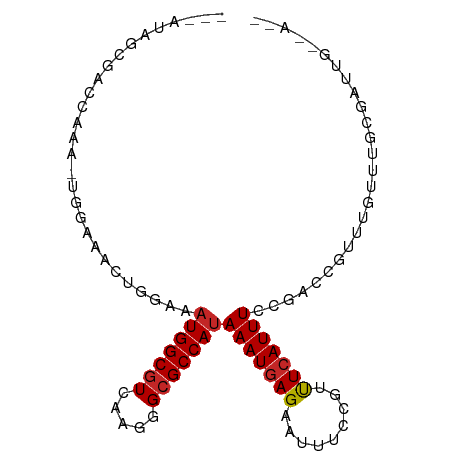
>dm3.chr2R 19028681 91 - 21146708 ---AUAGCGACCAAACUGGAAACUGGAAAUGGCGUCAAGGGCGCCAUAAAUGAGAAUUUCCGUUUCAUUUCCGACCGUUUGUUUGCGAUUGUGA-- ---...((((.(((((.(....)((((((((((((.....)))))))..((((((.......)))))))))))...))))).))))........-- ( -28.40, z-score = -2.01, R) >droSim1.chr2R 17610848 91 - 19596830 ---AUAGCGACCAAACUGGAAACUGGAAAUGGCGUCAAGGGCGCCAUAAAUGAGAAUUUCCGAUUCAUUUCCGACCGUUUGUUUGCGAUUGUGA-- ---...((((.(((((.(....)((((((((((((.....)))))........(((((...))))))))))))...))))).))))........-- ( -28.00, z-score = -2.02, R) >droSec1.super_9 2323222 84 - 3197100 ---AUAGCGACCAAA-------CUGGAAAUGGCGUCAAGGGCGCCAUAAAUGAGAAUUUCCGUUUCAUUUCCGACCGUUUGUUUGCGAUUGUGA-- ---...((((.((((-------(.((..(((((((.....)))))))((((((((.......))))))))....))))))).))))........-- ( -26.80, z-score = -2.23, R) >droEre2.scaffold_4845 20395820 91 - 22589142 ---AUAGCGACCAAACUGGAAACUGGAAAUGGCGUCAAGGGCGCCAUAAAUGAGAAUUUCCGUUUCAUUUCCGACCGUUUGUUUGCGAUUGUGA-- ---...((((.(((((.(....)((((((((((((.....)))))))..((((((.......)))))))))))...))))).))))........-- ( -28.40, z-score = -2.01, R) >droAna3.scaffold_13266 8126346 72 + 19884421 ---AUAGCGGCCGA--------CUGGAAAUGGCGUCAAGGGCGCCAUAAAUGAGAAUUUCCGUUUCAUUUCCGCCCGCUGGUU------------- ---.((((((.((.--------......(((((((.....)))))))((((((((.......)))))))).)).))))))...------------- ( -26.50, z-score = -1.81, R) >dp4.chr3 16507587 85 + 19779522 ---AUAGUGGGAAA--------CUGGAAAUGGCGUCAAGGGCGCCAUAAAUGAGAAUUUCCGUUUGAUUUCCGCCCGUUUGUUUGUGUGUGGCAGC ---.....((....--------))(((((((((((.....))))).((((((.(.....)))))))))))))(((((..(....)..)).)))... ( -23.80, z-score = -0.64, R) >droPer1.super_2 5571497 85 + 9036312 ---AUAGUGGGAAA--------CUGGAAAUGGCGUCAAGGGCGCCAUAAAUGAGAAUUUCCGUUUCAUUUCCGCCCGUUUGUUUGUGUGUGGCGGC ---.......((((--------(.(((((((((((.....)))))))..........)))))))))....(((((((..(....)..)).))))). ( -26.10, z-score = -1.00, R) >droVir3.scaffold_12875 2999653 89 - 20611582 CCAAUAGAUAGGAUAGUGGAAAUAGCGAAUGGCGUCAAGGGCGCCAUAAAUGAGAAUUUCCGACUCAUUUCCG-UUGUUUGCGUGUGUGA------ ((........))(((.((.((((((((.(((((((.....)))))))(((((((.........))))))).))-)))))).)))))....------ ( -25.20, z-score = -1.68, R) >droGri2.scaffold_15112 2016335 80 + 5172618 GAAAUAGAUAGGAUAGUGGAAAUAGCGAAUGGCGUCAAGGGCGCCAUAAAUGAGAAUUUCCGACUCAUUUCCG-UUGUUUG--------------- .((((((...(((.(((((((((..((.(((((((.....)))))))...))...)))))).)))....))).-)))))).--------------- ( -23.40, z-score = -2.64, R) >droMoj3.scaffold_6496 4670692 77 + 26866924 ---AUAGAGGGGAUAGUGGAAAUGGCGACUGGCAUCAAGGGCGCCAUAAAUGAGAAUUUCCGACUCAUUUCCG-UUGUUUG--------------- ---.......(((((((((((((((((.((.......))..))))))..(((((.........))))))))))-)))))).--------------- ( -20.30, z-score = -0.78, R) >consensus ___AUAGCGACCAAA_UGGAAACUGGAAAUGGCGUCAAGGGCGCCAUAAAUGAGAAUUUCCGUUUCAUUUCCGACCGUUUGUUUGCGAUUG__A__ ............................(((((((.....)))))))(((((((.........))))))).......................... (-12.50 = -12.59 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:55 2011