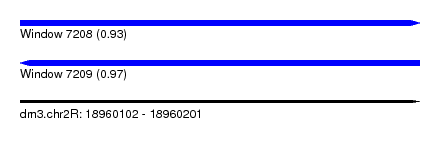
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,960,102 – 18,960,201 |
| Length | 99 |
| Max. P | 0.972748 |

| Location | 18,960,102 – 18,960,201 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.64 |
| Shannon entropy | 0.39499 |
| G+C content | 0.51917 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -32.31 |
| Energy contribution | -30.44 |
| Covariance contribution | -1.87 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.00 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.930376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
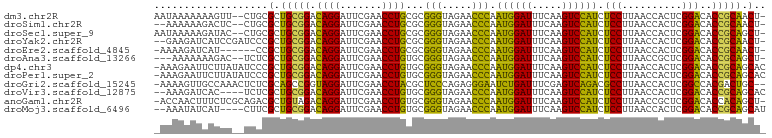
>dm3.chr2R 18960102 99 + 21146708 -AGUUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCGCAGGUUCGAAUCCUGUCCGCAGCGCAG--AACUUUUUUUAUU -.(((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......))))))))))))....--............. ( -29.80, z-score = -0.81, R) >droSim1.chr2R 17543678 97 + 19596830 -AGUUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCGCAGGUUCGAAUCCUGUCCGCAGCGCAG--GAGUCUUUUUU-- -.(((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......))))))))))))....--...........-- ( -29.80, z-score = -0.30, R) >droSec1.super_9 2251456 99 + 3197100 -AGCUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCGCAGGUUCGAAUCCUGUCCGCAGCGCAG--GUAUCUUUUUAUU -.(((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......))))))))))))....--............. ( -32.20, z-score = -0.98, R) >droYak2.chr2R 18922442 99 + 21139217 -AGUUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCGCAGGUUCGAAUCCUGUCCGCAGCGGGAUCGGAUGAUCUUC-- -.(((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......))))))))))))((((((....)))))).-- ( -33.70, z-score = -0.59, R) >droEre2.scaffold_4845 20323472 94 + 22589142 -AGUUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCGCAGGUUCGAAUCCUGUCCGCAGCGG------AUGAUCUUUU- -......((((((((.((((...((((..((((((((........((((.....))))..))))))))..))))..))))..)))------)).)))....- ( -31.70, z-score = -1.17, R) >droAna3.scaffold_13266 15289554 96 + 19884421 -AGCUGCGGUGUCCGAGCGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCGAAUCCUGUCCGCAGCGAGA--GUCUUUUUUU--- -.(((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......))))))))))))....--..........--- ( -33.80, z-score = -1.64, R) >dp4.chr3 16428222 101 - 19779522 GUGCUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCGAAUCCUGUCCGCAGCGGGAUAUAAGAAUUCUUU- ..(((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......))))))))))))(((((......)))))..- ( -32.70, z-score = -1.12, R) >droPer1.super_2 5491883 101 - 9036312 GUGCUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCGAAUCCUGUCCGCAGCGGGAUAUAAGAAUUCUUU- ..(((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......))))))))))))(((((......)))))..- ( -32.70, z-score = -1.12, R) >droGri2.scaffold_15245 3082834 99 - 18325388 --GCAGUCGUGGCCGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCGAGAGUUUGGCAACUUUU- --((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))(((((((....)))))))- ( -33.90, z-score = -0.71, R) >droVir3.scaffold_12875 11569921 96 - 20611582 GUGCUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCGAAUCCUGUCCGCAGCGAGA----GUGAUCUUU-- ..(((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......))))))))))))((((----....)))).-- ( -34.40, z-score = -1.76, R) >anoGam1.chr2R 51343480 100 - 62725911 -AGCUGUGGUGUCCGAGCGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCGAAUCCUGUCUACAGCGUCUGCGAGAAAGUUGGU- -.(((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......))))))))))))..................- ( -30.00, z-score = -0.13, R) >droMoj3.scaffold_6496 18131756 96 + 26866924 AUGCUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCGAAUCCUGUCCGCAGCGAAG----AUGAUAUUU-- .((((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......)))))))))))))...----.........-- ( -32.00, z-score = -1.64, R) >consensus _AGCUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCGAAUCCUGUCCGCAGCGAGA__GAAUGAUUUUUU_ ..(((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......))))))))))))................... (-32.31 = -30.44 + -1.87)

| Location | 18,960,102 – 18,960,201 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.64 |
| Shannon entropy | 0.39499 |
| G+C content | 0.51917 |
| Mean single sequence MFE | -28.39 |
| Consensus MFE | -22.69 |
| Energy contribution | -21.21 |
| Covariance contribution | -1.48 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.972748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18960102 99 - 21146708 AAUAAAAAAAGUU--CUGCGCUGCGGACAGGAUUCGAACCUGCGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAACU- ..........(((--((((.(((((..((((.......))))))))))))))))...((((((.....))))))........((....))...........- ( -26.80, z-score = -1.61, R) >droSim1.chr2R 17543678 97 - 19596830 --AAAAAAGACUC--CUGCGCUGCGGACAGGAUUCGAACCUGCGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAACU- --......((.((--((((......).))))).)).....((((.(((.....))).((((((.....))))))........((....))....))))...- ( -23.20, z-score = -0.39, R) >droSec1.super_9 2251456 99 - 3197100 AAUAAAAAGAUAC--CUGCGCUGCGGACAGGAUUCGAACCUGCGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAGCU- .............--....(((((((...((.(((..((((....)))).))).)).((((((.....))))))........((....))...))))))).- ( -29.60, z-score = -2.23, R) >droYak2.chr2R 18922442 99 - 21139217 --GAAGAUCAUCCGAUCCCGCUGCGGACAGGAUUCGAACCUGCGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAACU- --...((((....))))....(((((...((.(((..((((....)))).))).)).((((((.....))))))........((....))...)))))...- ( -25.80, z-score = -0.62, R) >droEre2.scaffold_4845 20323472 94 - 22589142 -AAAAGAUCAU------CCGCUGCGGACAGGAUUCGAACCUGCGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAACU- -..........------....(((((...((.(((..((((....)))).))).)).((((((.....))))))........((....))...)))))...- ( -23.00, z-score = -0.55, R) >droAna3.scaffold_13266 15289554 96 - 19884421 ---AAAAAAAGAC--UCUCGCUGCGGACAGGAUUCGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCGCUCGGACACCGCAGCU- ---..........--....((((((((((((.......)))))(((((.....))).((((((.....))))))..........)).......))))))).- ( -32.80, z-score = -2.92, R) >dp4.chr3 16428222 101 + 19779522 -AAAGAAUUCUUAUAUCCCGCUGCGGACAGGAUUCGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAGCAC -..................((((((((((((.......)))))..(((.....))).((((((.....))))))........((....))...))))))).. ( -31.30, z-score = -2.79, R) >droPer1.super_2 5491883 101 + 9036312 -AAAGAAUUCUUAUAUCCCGCUGCGGACAGGAUUCGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAGCAC -..................((((((((((((.......)))))..(((.....))).((((((.....))))))........((....))...))))))).. ( -31.30, z-score = -2.79, R) >droGri2.scaffold_15245 3082834 99 + 18325388 -AAAAGUUGCCAAACUCUCGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCGGCCACGACUGC-- -....((.(((...((((.(.(((..(((((.......)))))))).).))))((..((((((.....))))))..))..........))).))......-- ( -29.50, z-score = -1.38, R) >droVir3.scaffold_12875 11569921 96 + 20611582 --AAAGAUCAC----UCUCGCUGCGGACAGGAUUCGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAGCAC --.........----....((((((((((((.......)))))..(((.....))).((((((.....))))))........((....))...))))))).. ( -31.30, z-score = -2.71, R) >anoGam1.chr2R 51343480 100 + 62725911 -ACCAACUUUCUCGCAGACGCUGUAGACAGGAUUCGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCGCUCGGACACCACAGCU- -..................(((((.((((((.......)))))(((((.....))).((((((.....))))))..........)).......).))))).- ( -24.80, z-score = -0.59, R) >droMoj3.scaffold_6496 18131756 96 - 26866924 --AAAUAUCAU----CUUCGCUGCGGACAGGAUUCGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAGCAU --.........----....((((((((((((.......)))))..(((.....))).((((((.....))))))........((....))...))))))).. ( -31.30, z-score = -3.14, R) >consensus _AAAAAAUCAUUC__UCCCGCUGCGGACAGGAUUCGAACCUGCGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAGCU_ ...................(.(((((.((((.......))))...(((.....))).((((((.....)))))).(((..........)))..))))).).. (-22.69 = -21.21 + -1.48)
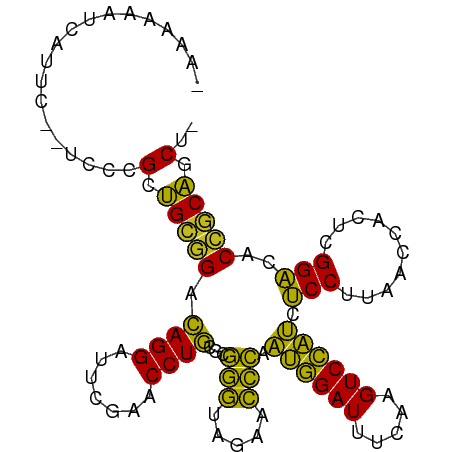
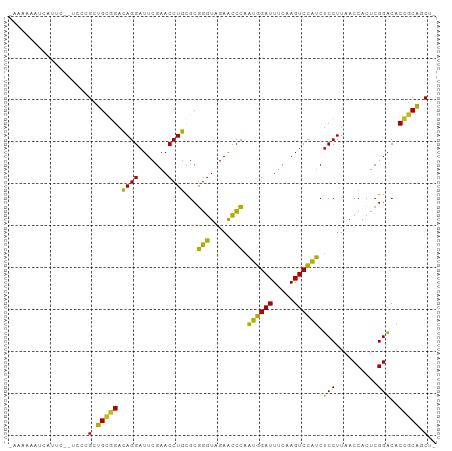
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:49 2011