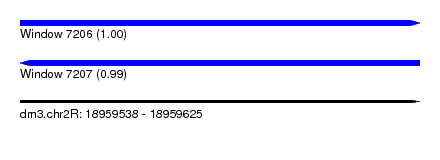
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,959,538 – 18,959,625 |
| Length | 87 |
| Max. P | 0.998484 |

| Location | 18,959,538 – 18,959,625 |
|---|---|
| Length | 87 |
| Sequences | 13 |
| Columns | 87 |
| Reading direction | forward |
| Mean pairwise identity | 89.68 |
| Shannon entropy | 0.24222 |
| G+C content | 0.54616 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -30.63 |
| Energy contribution | -29.11 |
| Covariance contribution | -1.52 |
| Combinations/Pair | 1.50 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.37 |
| SVM RNA-class probability | 0.998484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
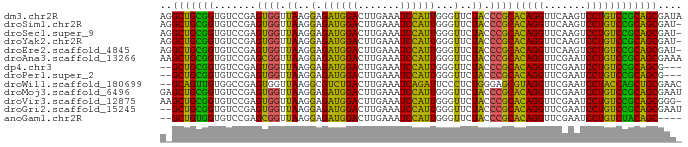
>dm3.chr2R 18959538 87 + 21146708 UAUCGCUGCGGACAGGACUUGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAGCCU ....(((((((...(((((((((.((((..(((.....))).))))..)))))))))..........((....))...))))))).. ( -35.50, z-score = -4.62, R) >droSim1.chr2R 17543106 86 + 19596830 -AUCGCUGCGGACAGGACUUGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAGCCU -...(((((((...(((((((((.((((..(((.....))).))))..)))))))))..........((....))...))))))).. ( -35.50, z-score = -4.51, R) >droSec1.super_9 2250884 86 + 3197100 -AUCGCUGCGGACAGGACUUGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAGCCU -...(((((((...(((((((((.((((..(((.....))).))))..)))))))))..........((....))...))))))).. ( -35.50, z-score = -4.51, R) >droYak2.chr2R 18921864 86 + 21139217 -AUCGCUGCGGACAGGACUUGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAGCCU -...(((((((...(((((((((.((((..(((.....))).))))..)))))))))..........((....))...))))))).. ( -35.50, z-score = -4.51, R) >droEre2.scaffold_4845 20322904 86 + 22589142 -AUCGCUGCGGACAGGACUUGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAGCCU -...(((((((...(((((((((.((((..(((.....))).))))..)))))))))..........((....))...))))))).. ( -35.50, z-score = -4.51, R) >droAna3.scaffold_13266 15288896 87 + 19884421 UUUCGCUGCGGACAGGAUUCGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCGCUCGGACACCGCAGCUU ....((((((((((((.......)))))(((((.....))).((((((.....))))))..........)).......))))))).. ( -32.80, z-score = -3.21, R) >dp4.chr3 16427764 82 - 19779522 ---CGCUGCGGACAGGAUUCGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAGC-- ---.((((((((((((.......)))))..(((.....))).((((((.....))))))........((....))...)))))))-- ( -29.60, z-score = -2.80, R) >droPer1.super_2 5491427 82 - 9036312 ---CGCUGCGGACAGGAUUCGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAGC-- ---.((((((((((((.......)))))..(((.....))).((((((.....))))))........((....))...)))))))-- ( -29.60, z-score = -2.80, R) >droWil1.scaffold_180699 2076854 85 - 2593675 GUUCGCAGCUGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCAAGUCAGACGCCUUAACCACUCGGCCACAACUGC-- ....((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)).))))-- ( -24.82, z-score = -1.39, R) >droMoj3.scaffold_6496 18131508 87 + 26866924 AUUCGCUGCGGACAGGAUUCGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAGCUC ....((((((((((((.......)))))..(((.....))).((((((.....))))))........((....))...))))))).. ( -30.80, z-score = -2.94, R) >droVir3.scaffold_12875 11569674 86 - 20611582 -CCCGCUGCGGACAGGAUUCGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAGCUU -...((((((((((((.......)))))..(((.....))).((((((.....))))))........((....))...))))))).. ( -30.80, z-score = -2.92, R) >droGri2.scaffold_15245 15949272 85 - 18325388 AUUCGCUGCGGACAGGAUUCGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAGC-- ....((((((((((((.......)))))..(((.....))).((((((.....))))))........((....))...)))))))-- ( -29.60, z-score = -2.74, R) >anoGam1.chr2R 51289722 81 - 62725911 ----GCUGUAGACAGGAUUCGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCGCUCGGACACCACAGC-- ----(((((.((((((.......)))))(((((.....))).((((((.....))))))..........)).......).)))))-- ( -23.30, z-score = -1.40, R) >consensus _AUCGCUGCGGACAGGAUUCGAACCUGUGCGGGUAGAACCCAAUGGAUUUCAAGUCCAUCUCCUUAACCACUCGGACACCGCAGC_U ....((((((((((((.......)))))..(((.....))).((((((.....)))))).(((..........)))..))))))).. (-30.63 = -29.11 + -1.52)


| Location | 18,959,538 – 18,959,625 |
|---|---|
| Length | 87 |
| Sequences | 13 |
| Columns | 87 |
| Reading direction | reverse |
| Mean pairwise identity | 89.68 |
| Shannon entropy | 0.24222 |
| G+C content | 0.54616 |
| Mean single sequence MFE | -32.91 |
| Consensus MFE | -32.25 |
| Energy contribution | -30.88 |
| Covariance contribution | -1.37 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.993871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
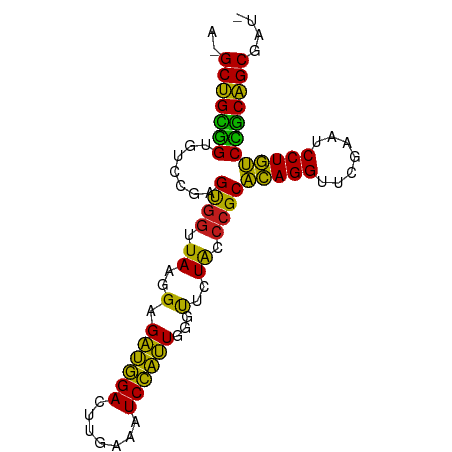
>dm3.chr2R 18959538 87 - 21146708 AGGCUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCAAGUCCUGUCCGCAGCGAUA ..((((((((.(((..........))).).(((((((((......((((.....))))......)))))))))...))))))).... ( -35.40, z-score = -2.81, R) >droSim1.chr2R 17543106 86 - 19596830 AGGCUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCAAGUCCUGUCCGCAGCGAU- ..((((((((.(((..........))).).(((((((((......((((.....))))......)))))))))...)))))))...- ( -35.40, z-score = -2.84, R) >droSec1.super_9 2250884 86 - 3197100 AGGCUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCAAGUCCUGUCCGCAGCGAU- ..((((((((.(((..........))).).(((((((((......((((.....))))......)))))))))...)))))))...- ( -35.40, z-score = -2.84, R) >droYak2.chr2R 18921864 86 - 21139217 AGGCUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCAAGUCCUGUCCGCAGCGAU- ..((((((((.(((..........))).).(((((((((......((((.....))))......)))))))))...)))))))...- ( -35.40, z-score = -2.84, R) >droEre2.scaffold_4845 20322904 86 - 22589142 AGGCUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCAAGUCCUGUCCGCAGCGAU- ..((((((((.(((..........))).).(((((((((......((((.....))))......)))))))))...)))))))...- ( -35.40, z-score = -2.84, R) >droAna3.scaffold_13266 15288896 87 - 19884421 AAGCUGCGGUGUCCGAGCGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCGAAUCCUGUCCGCAGCGAAA ..(((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......)))))))))))).... ( -33.80, z-score = -2.43, R) >dp4.chr3 16427764 82 + 19779522 --GCUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCGAAUCCUGUCCGCAGCG--- --(((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......)))))))))))).--- ( -31.70, z-score = -2.29, R) >droPer1.super_2 5491427 82 + 9036312 --GCUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCGAAUCCUGUCCGCAGCG--- --(((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......)))))))))))).--- ( -31.70, z-score = -2.29, R) >droWil1.scaffold_180699 2076854 85 + 2593675 --GCAGUUGUGGCCGAGUGGUUAAGGCGUCUGACUUGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCAGCUGCGAAC --((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))).... ( -30.00, z-score = -1.75, R) >droMoj3.scaffold_6496 18131508 87 - 26866924 GAGCUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCGAAUCCUGUCCGCAGCGAAU ..(((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......)))))))))))).... ( -31.90, z-score = -1.94, R) >droVir3.scaffold_12875 11569674 86 + 20611582 AAGCUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCGAAUCCUGUCCGCAGCGGG- ..(((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......))))))))))))...- ( -31.90, z-score = -1.62, R) >droGri2.scaffold_15245 15949272 85 + 18325388 --GCUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCGAAUCCUGUCCGCAGCGAAU --(((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......)))))))))))).... ( -31.70, z-score = -2.12, R) >anoGam1.chr2R 51289722 81 + 62725911 --GCUGUGGUGUCCGAGCGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCGAAUCCUGUCUACAGC---- --(((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......))))))))))))---- ( -28.10, z-score = -1.69, R) >consensus A_GCUGCGGUGUCCGAGUGGUUAAGGAGAUGGACUUGAAAUCCAUUGGGUUCUACCCGCACAGGUUCGAAUCCUGUCCGCAGCGAU_ ..(((((((.......((((.((..(.((((((.......))))))...)..)).))))(((((.......)))))))))))).... (-32.25 = -30.88 + -1.37)
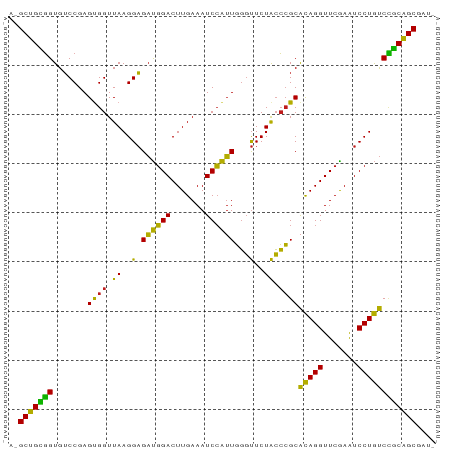
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:47 2011