
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,921,072 – 18,921,168 |
| Length | 96 |
| Max. P | 0.506203 |

| Location | 18,921,072 – 18,921,168 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 60.68 |
| Shannon entropy | 0.80223 |
| G+C content | 0.43702 |
| Mean single sequence MFE | -18.46 |
| Consensus MFE | -6.94 |
| Energy contribution | -6.76 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.18 |
| Mean z-score | -0.67 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.506203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
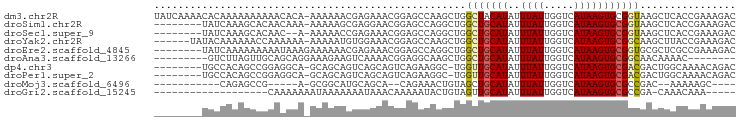
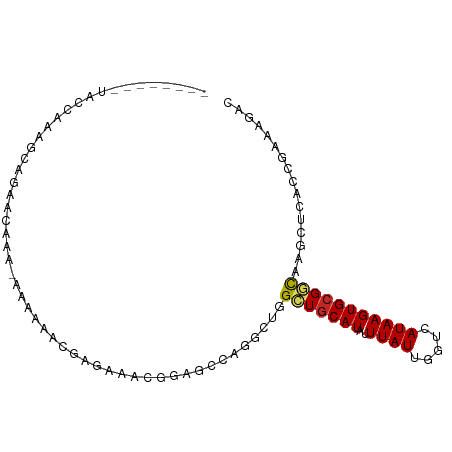
>dm3.chr2R 18921072 96 - 21146708 UAUCAAAACACAAAAAAAAAACACA-AAAAAACGAGAAACGGAGCCAAGCUGGCUACAUAUUUAUUGGUCAUAAGUGCGGUAAGCUCACCGAAAGAC .........................-.......(((..((.(((((.....))))...(((((((.....)))))))).))...)))..(....).. ( -12.60, z-score = -0.28, R) >droSim1.chr2R 17509347 88 - 19596830 --------UAUCAAAGCACAACAAA-AAAAAGCGAGGAACGGAGCCAGGCUGGCUGCAUAUUUAUUGGUCAUAAGUGCGGUAAGCUCACCGAAAGAC --------..((...((........-.....))...)).(((.(...((((.(((((((..((((.....))))))))))).))))).)))...... ( -17.32, z-score = 0.07, R) >droSec1.super_9 2212726 86 - 3197100 --------UAUCAAAGCACAAC--A-AAAAACCGAGAAACGGAGCCAGGCUGGCUGCAUAUUUAUUGGUCAUAAGUGCGGUAAGCUCACCGAAAGAC --------..((...((((...--.-....(((((.(((.(.((((.....)))).)...))).))))).....))))(((......)))))..... ( -18.32, z-score = -0.70, R) >droYak2.chr2R 18882697 90 - 21139217 ------UAUACAAAAAACCAAAAAA-AAAAAUGUGGAAACGGAGCCAAGCUGGCUGCAUAUUUAUUGGUCAUAAGUGCGGCAAGCUUACCGAAAGAC ------...........(((.....-.......)))...(((....(((((.(((((((..((((.....))))))))))).))))).)))...... ( -19.10, z-score = -1.29, R) >droEre2.scaffold_4845 20283909 89 - 22589142 --------UAUCAAAAAAAAAUAAAGAAAAAACGAGAAACGGAGCCAGGCUGGCUGCAUAUUUAUUGGUCAUAAGUGCGGUGCGCUCGCCGAAAGAC --------........................((((..(((.((((.....)))).).(((((((.....)))))))..))...)))).(....).. ( -16.40, z-score = 0.30, R) >droAna3.scaffold_13266 15254819 80 - 19884421 ---------GUCUUAGUUGCAGCAGGAAAGAAGUCAAAACGGAGGCAAGCUGGCUGCAUAUUUAUUGGUCAUAAGUGCGGCAACAAAAC-------- ---------......(((((.((((...((..(((........)))...))..)))).(((((((.....)))))))..))))).....-------- ( -17.10, z-score = -0.05, R) >dp4.chr3 2458594 87 - 19779522 --------UGCCACAGCCGGAGGCA-GCAGCAGUCAGCAGUCAGAAGGC-UGGUUGCAUAUUUAUUGGUCAUAAGUGCGACGACUGGCAAAACAGAC --------((((((((((...(((.-((........)).)))....)))-))(((((((..((((.....)))))))))))...)))))........ ( -27.50, z-score = -1.54, R) >droPer1.super_2 2647363 87 - 9036312 --------UGCCACAGCCGGAGGCA-GCAGCAGUCAGCAGUCAGAAGGC-UGGUUGCAUAUUUAUUGGUCAUAAGUGCGACGACUGGCAAAACAGAC --------((((((((((...(((.-((........)).)))....)))-))(((((((..((((.....)))))))))))...)))))........ ( -27.50, z-score = -1.54, R) >droMoj3.scaffold_6496 25806927 72 - 26866924 -----------CAGAGCCG-----A-GCGGCAUGCAGCA--CAGAAACUGUAGCUGCAUAUUUAUUGGUCAUAAGUGCGCCGAC--AAAAAGC---- -----------....(((.-----.-..)))((((((((--(((...)))).))))))).(((.(((((((....)).))))).--)))....---- ( -22.70, z-score = -1.91, R) >droGri2.scaffold_15245 641170 72 - 18325388 -------------------CAAAAAAAUAAAAAAAUAAACAAAAAUACUGUAGUUGCAUAUUUAUUGGUCAUAAGUGCGCCGA-CAAACAAA----- -------------------.................................(((((.(((((((.....))))))).).)))-).......----- ( -6.10, z-score = 0.22, R) >consensus ________UACCAAAGCAGAACAAA_AAAAAACGAGAAACGGAGCCAGGCUGGCUGCAUAUUUAUUGGUCAUAAGUGCGGCAAGCUCACCGAAAGAC ....................................................(((((((..((((.....)))))))))))................ ( -6.94 = -6.76 + -0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:42 2011