| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,901,312 – 18,901,420 |
| Length | 108 |
| Max. P | 0.540015 |

| Location | 18,901,312 – 18,901,420 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.03 |
| Shannon entropy | 0.49992 |
| G+C content | 0.52680 |
| Mean single sequence MFE | -30.29 |
| Consensus MFE | -17.68 |
| Energy contribution | -19.39 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.15 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.540015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
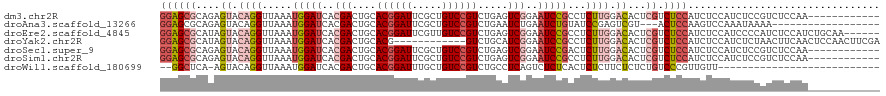
>dm3.chr2R 18901312 108 - 21146708 GGAGCGCAGAGUACAGGUUAAAUGGAUCACGACUGCACGGAUUCGCUGUCCGUCUGAGUCGGAAUCCGCCUCUUGGACACUCGUCUCCAUCUCCAUCUCCGUCUCCAA------------ ((((((.(((...((((.....(((((..(((((..((((((.....))))))...)))))..)))))...))))(((....)))..........))).)).))))..------------ ( -36.70, z-score = -1.68, R) >droAna3.scaffold_13266 15237278 99 - 19884421 GGAGCGCAGAGUACAGGUUAAAUGGAUCACGACUGCACGGAUUCGCUGUCCGUCUGAAUCUGAAUCUGUAUCCGAGUCGU---ACUCCAAGUCCAAAUAAAA------------------ (((..(((((...((((((..((((((..(((((....))..)))..))))))...))))))..))))).)))(((....---.)))...............------------------ ( -27.30, z-score = -1.21, R) >droEre2.scaffold_4845 20263023 114 - 22589142 GGAGCGCAUAGUACAGGUUAAAUGGAUCACGACUGCACGGAUUCGUUGUCCGUCUGAGUCGGAAUCCGCCUCUUGGACACUCGUCUCCAUCUCCAUCCCCAUCUCCAUCUGCAA------ ((((((....((.((((.....(((((..(((((..((((((.....))))))...)))))..)))))...)))).))...)).))))..........................------ ( -32.20, z-score = -0.76, R) >droYak2.chr2R 18862214 108 - 21139217 GGAGCGCAUAGUACAGGUUAAAUGGAUCACGACUGCACG------------GUCUGCAUCGGAAUCCGCCUCUUGGACACUCGUCUCCAUCUCCAUCUCUAACUUCAACUCCAACUUCGA ((((.....(((.((((.....(((((..(((.((((..------------...)))))))..)))))...))))(((....)))................)))....))))........ ( -24.60, z-score = -0.59, R) >droSec1.super_9 2193179 108 - 3197100 GGAGCGCAGAGUACAGGUUAAAUGGAUCACGACUGCACGGAUUCGCUGUCCGUCUGAGUCGGAAUCCGACUCUUGGACACUCGUCUCCAUCUCCAUCUCCGUCUCCAA------------ ((((.((.(((....((....(((((..((((..((........))((((((...(((((((...))))))).)))))).)))).)))))..))..))).))))))..------------ ( -38.90, z-score = -2.21, R) >droSim1.chr2R 17489664 108 - 19596830 GGAGCGCAGAGUACAGGUUAAAUGGAUCACGACUGCACGGAUUCGCUGUCCGUCUGAGUCGGAAUCCGCCUCUUGGACACUCGUCUCCAUCUCCAUCUCCGUCUCCAA------------ ((((((.(((...((((.....(((((..(((((..((((((.....))))))...)))))..)))))...))))(((....)))..........))).)).))))..------------ ( -36.70, z-score = -1.68, R) >droWil1.scaffold_180699 1980550 90 + 2593675 --GGCUCA-AGUACAGGUUAAAUGGAUCACGACUGCACGGAUUUGCUGUCCGUCUGCCUCAGUCUCUCACUCUCUUCUCUCUGUCCCGUUGUU--------------------------- --((..((-......((((.....))))..(((((.((((((.....))))))......))))).................))..))......--------------------------- ( -15.60, z-score = 1.24, R) >consensus GGAGCGCAGAGUACAGGUUAAAUGGAUCACGACUGCACGGAUUCGCUGUCCGUCUGAGUCGGAAUCCGCCUCUUGGACACUCGUCUCCAUCUCCAUCUCCAUCUCCAA____________ ((((((....((.((((.....(((((..(((....((((((.....)))))).....)))..)))))...)))).))...)).))))................................ (-17.68 = -19.39 + 1.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:38 2011