
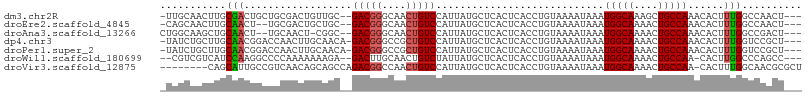
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,896,258 – 18,896,359 |
| Length | 101 |
| Max. P | 0.715249 |

| Location | 18,896,258 – 18,896,359 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.18 |
| Shannon entropy | 0.46641 |
| G+C content | 0.48290 |
| Mean single sequence MFE | -25.56 |
| Consensus MFE | -11.93 |
| Energy contribution | -12.01 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.715249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
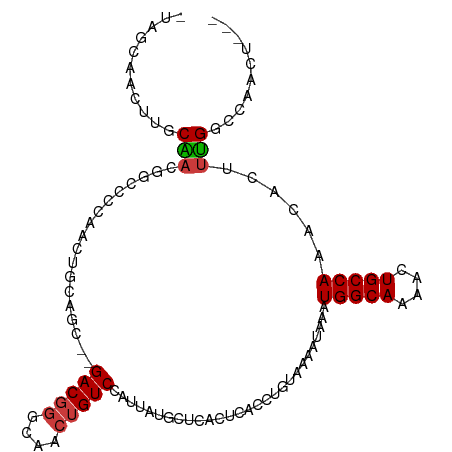
>dm3.chr2R 18896258 101 + 21146708 -UUGCAACUUGCGACUGCUGCGACUGUUGC--GACGGGCAACUGUCCAUUAUGCUCACUCACCUGUAAAAUAAAUGGCAAAGCUGCCAAACACUUUGGCCAACU--- -..((((((((((.....)))))..)))))--(((((....)))))............................((((((((.((.....)))))).))))...--- ( -25.50, z-score = -0.96, R) >droEre2.scaffold_4845 20257914 99 + 22589142 -CAGCAACUUGCAACU--UGCGACUGCUGC--GACGGGCAACUGUCCAUUAUGCUCACUCACCUGUAAAAUAAAUGGCAAAACUGCCAAACACUUUGGCCAACU--- -(((((..((((....--.)))).))))).--(((((....)))))......(((.((......))........(((((....)))))........))).....--- ( -24.40, z-score = -1.79, R) >droAna3.scaffold_13266 15230781 99 + 19884421 CUGGCAAGCUGCAACU--UGCAACU-CGGC--GACGGGCAACUGUCCAUUAUGCUCACUCACCUGUAAAAUAAAUGGCAAAACUGCCAAACACUUUGGCCGACU--- ...(((((......))--)))...(-((((--(((((....)))))............................(((((....))))).........)))))..--- ( -29.50, z-score = -2.94, R) >dp4.chr3 2431115 102 + 19779522 -UAUCUGCUUGCAACGGACCAACUUGCAACA-GACGGGCCGCUGUCCAUUAUGCUCACUCACCUGUAAAAUAAAUGGCAAAACUGCCAAACACUUUGGUCCGCU--- -..((((.((((((.(......)))))))))-))(((((((..((..(((.(((..........))).)))...(((((....)))))...))..)))))))..--- ( -28.10, z-score = -2.58, R) >droPer1.super_2 2620877 102 + 9036312 -UAUCUGCUUGCAACGGACCAACUUGCAACA-GACGGGCCGCUGUCCAUUAUGCUCACUCACCUGUAAAAUAAAUGGCAAAACUGCCAAACACUUUGGUCCGCU--- -..((((.((((((.(......)))))))))-))(((((((..((..(((.(((..........))).)))...(((((....)))))...))..)))))))..--- ( -28.10, z-score = -2.58, R) >droWil1.scaffold_180699 1973876 99 - 2593675 --CGUCGUCAUCCAAGGCCCCAAAAAAAGA--GACUUGCAACUGUCUAUUAUGCUCACUCACCUGUAAAAUAAAUGGCAAAACUGCCAA-CACUUGGCCCAGCC--- --.............((((..........(--(((........))))...........................(((((....))))).-.....)))).....--- ( -17.30, z-score = -0.31, R) >droVir3.scaffold_12875 7066577 98 + 20611582 --------CAGCAUUGCCGUCAACAGCAGCCAGACGGCCAACUGUCCAUUAUGCUCACUCACCUGUAAAAUAAAUGGCAAAACUGCCAA-CACUUUGGCAACGCGCU --------..((.((((((.....((((....(((((....))))).....))))...................(((((....))))).-.....)))))).))... ( -26.00, z-score = -1.46, R) >consensus _UAGCAACUUGCAACGGCCCCAACUGCAGC__GACGGGCAACUGUCCAUUAUGCUCACUCACCUGUAAAAUAAAUGGCAAAACUGCCAAACACUUUGGCCAACU___ ...........(((..................(((((....)))))............................(((((....)))))......))).......... (-11.93 = -12.01 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:36 2011