
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,891,126 – 18,891,222 |
| Length | 96 |
| Max. P | 0.834625 |

| Location | 18,891,126 – 18,891,222 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 72.49 |
| Shannon entropy | 0.55295 |
| G+C content | 0.52890 |
| Mean single sequence MFE | -21.53 |
| Consensus MFE | -13.87 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
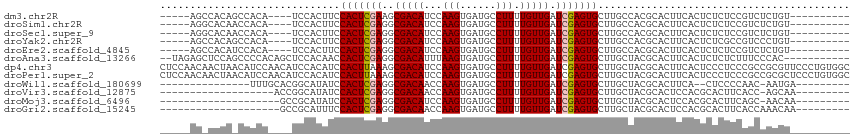
>dm3.chr2R 18891126 96 - 21146708 -----AGCCACAGCCACA----UCCACUUCCACUCGAAGCGACAUCCAAGUGAUGCCUUUUGUUGAUCGAGUGCUUGCCACGCACUUCACUCUCUCCGUCUCUGU---------- -----....((((..((.----........(((((((..(((((...(((......))).))))).)))))))..(((...))).............))..))))---------- ( -18.00, z-score = 0.17, R) >droSim1.chr2R 17479019 96 - 19596830 -----AGGCACAACCACA----UCCACUUCCACUCGAGGCGACAUCCAAGUGAUGCCUUUUGUUGAUCGAGUGCUUGCCACGCACUUCACUCUCUCCGUCUCUGU---------- -----((((.........----......((.((..((((((.(((....))).))))))..)).))..(((((..(((...)))...))))).....))))....---------- ( -21.30, z-score = -0.28, R) >droSec1.super_9 2182953 96 - 3197100 -----AGGCACAACCACA----UCCACUUCCACUCGAGGCGACAUCCAAGUGAUGCCUUUUGUUGAUCGAGUGCUUGCCACGCACUUCACUCUCUCCGUCUCUGU---------- -----((((.........----......((.((..((((((.(((....))).))))))..)).))..(((((..(((...)))...))))).....))))....---------- ( -21.30, z-score = -0.28, R) >droYak2.chr2R 18851644 96 - 21139217 -----AGCCACAGCCACA----UCCACUUCCACUCGAGGCGACAUCCAAGUGAUGCCUUUUGUUGAUCGAGUGCUUGCCACGCACUUCACUCUCGCCGUCCCUGU---------- -----....((((..((.----......((.((..((((((.(((....))).))))))..)).))..(((((..(((...)))...))))).....))..))))---------- ( -22.10, z-score = -0.41, R) >droEre2.scaffold_4845 20252712 96 - 22589142 -----AGCCACAUCCACA----UCCACUUCCACUCGAGGCGACAUCCAAGUGAUGCCUUUUGUUGAUCGAGUGCUUGCCACGCACUUCACUCUCUCCGUCUCUGU---------- -----....(((...((.----......((.((..((((((.(((....))).))))))..)).))..(((((..(((...)))...))))).....))...)))---------- ( -18.50, z-score = 0.11, R) >droAna3.scaffold_13266 15225929 102 - 19884421 --UAGAGCUCCAGCCCCACAGCUCCACAACCACUCGAGGCGACAUUUAAGUGAUGCCUUUUGUUGAUCGAGUGCUUGCUACGCACUUCACUCUCUCUUUCCCAC----------- --..(((((..........)))))..((((.....((((((.(((....))).))))))..))))...(((((..(((...)))...)))))............----------- ( -23.00, z-score = -1.31, R) >dp4.chr3 2426279 115 - 19779522 CUCCAACAACUAACAUCCAACAUCCACAUCCACUUAAAGCGACAUCCAAGUGAUGCCUUUUGUUGAUCGAGUGCUUGCUACGCACUUCACUCCCUCCCGCCGCGUUCCCUGUGGC .................(((((....((((.((((............)))))))).....)))))...(((((..(((...)))...)))))......((((((.....)))))) ( -22.20, z-score = -1.53, R) >droPer1.super_2 2615981 115 - 9036312 CUCCAACAACUAACAUCCAACAUCCACAUCCACUUAAAGCGACAUCCAAGUGAUGCCUUUUGUUGAUCGAGUGCUUGCUACGCACUUCACUCCCUCCCGCCGCGCUCCCUGUGGC .................(((((....((((.((((............)))))))).....)))))...(((((..(((...)))...)))))......((((((.....)))))) ( -22.10, z-score = -1.49, R) >droWil1.scaffold_180699 1965793 88 + 2593675 ---------------UUUGCACGGCAUAUCCACUCGAGGCGACAACCAAGUGAUGCCUUUUGUUGAUCGAGUGCUUGCUACGCACUUCA--CUCCCCAAC-AAUGA--------- ---------------..(((..((((....(((((((..((((((..(((......))))))))).)))))))..))))..))).....--.........-.....--------- ( -23.00, z-score = -1.70, R) >droVir3.scaffold_12875 7061417 86 - 20611582 -------------------ACCGGCAUAUCCACUCGAGGCGACAACCAAGUGAUGCCUUUUGUUGAUCGAGUGCUUGCUACGCACUCCACGCACUUCACC-AGCAA--------- -------------------....((..(((.((..((((((.((......)).))))))..)).))).((((((.......))))))...))........-.....--------- ( -21.90, z-score = -0.97, R) >droMoj3.scaffold_6496 25767420 85 - 26866924 --------------------GCCGCAUAUCCACUCGAGGCGACAUCCAAGUGAUGCCUUUUGUUGAUCGAGUGCUUGCUACGCACUCCACGCACUUCAGC-AACAA--------- --------------------...((..(((.((..((((((.(((....))).))))))..)).))).((((((.......))))))...))........-.....--------- ( -23.30, z-score = -1.44, R) >droGri2.scaffold_15245 608163 86 - 18325388 --------------------GCCGCAUUUCCACUCGAGGCGACAACCAAGUGAUGCCUUUUGUUGAUCGAGUGCUUGCUACGCACUCCACGCACUUCACCAAACAA--------- --------------------...((...((.((..((((((.((......)).))))))..)).))..((((((.......))))))...))..............--------- ( -21.60, z-score = -1.61, R) >consensus _____AG_CACA_CCACA____UCCACAUCCACUCGAGGCGACAUCCAAGUGAUGCCUUUUGUUGAUCGAGUGCUUGCUACGCACUUCACUCUCUCCGUCCCUGU__________ ..............................(((((((..(((((...(((......))).))))).))))))).......................................... (-13.87 = -13.90 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:35 2011