
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,843,461 – 18,843,571 |
| Length | 110 |
| Max. P | 0.843238 |

| Location | 18,843,461 – 18,843,571 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 73.55 |
| Shannon entropy | 0.54732 |
| G+C content | 0.54618 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -23.18 |
| Energy contribution | -25.20 |
| Covariance contribution | 2.02 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.843238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
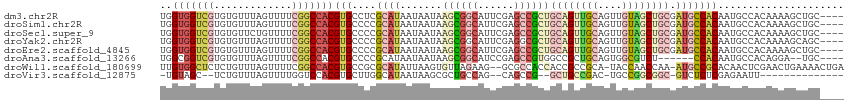
>dm3.chr2R 18843461 110 - 21146708 UGGUGGUCGUGUGUUUAGUUUUCGGCCACGUGCCUCGCAUAAUAAUAAGCGGCAUUCGAGCCGCUGCAGUUGCAGUUGUAGCUGCGAUGCCACAAUGCCACAAAAGCUGC---- ..(((((..((((..........(((.....)))..((((.......((((((......))))))((((((((....)))))))).))))))))..))))).........---- ( -42.70, z-score = -1.82, R) >droSim1.chr2R 17438425 110 - 19596830 UGGUGGUCGUGUGUUUAGUUUUCGGCCACGUGCCCCGCAUAAUAAUAAGCGGCAUUCGAGCCGCUGCAGUUGCAGUUGUAGCUGCGAUGCCACAAUGCCACAAAAGCUGC---- ..(((((..((((..........(((.....)))..((((.......((((((......))))))((((((((....)))))))).))))))))..))))).........---- ( -42.30, z-score = -1.76, R) >droSec1.super_9 2140662 110 - 3197100 UGGUGGUCGUGUGUUCUGUUUUCGGCCACGUGCCCCGCAUAAUAAUAAGCGGCAUUCGAGCCGCUGCAGUUGCAGUUGUAGCUGCGAUGCCACAAUGCCACAAAAGCUGC---- ..(((((..((((..........(((.....)))..((((.......((((((......))))))((((((((....)))))))).))))))))..))))).........---- ( -42.30, z-score = -1.61, R) >droYak2.chr2R 18808347 110 - 21139217 UGGUGGUCGUGUGUUUAGUUUUCGGCCACGUGCCCCGCAUAAUAAUAAGCGGCAUUCGAGGCGCUGCAGUUGCAGUUGUAGCUGCGAUGCCACAAUGCCACAAAAGCAGC---- ..(((((((.............))))))).(((...((..........))((((((.(.((((.(((((((((....))))))))).)))).)))))))......)))..---- ( -42.12, z-score = -1.53, R) >droEre2.scaffold_4845 20205536 110 - 22589142 UGGUGGUCGUGUGUUUAGUUUUCGGCCACGUGCCCCGCAUAAUAAUAAGCGGCAUUCGAGCCGCUGCAGUUGCAGUUGUAGCUGCGAUGCCACAAUGCCACAAAAGCUGC---- ..(((((..((((..........(((.....)))..((((.......((((((......))))))((((((((....)))))))).))))))))..))))).........---- ( -42.30, z-score = -1.76, R) >droAna3.scaffold_13266 15181802 102 - 19884421 UGGCGGUCGUGUGUUUAGUUUUCGGCCACGUGCCCCGCAUAAUAAUAAGCGGCAUCCGAGCCGUGGCCGCUGCAGUGGCGUCU------CCACAAUGCCACAGGA--UGC---- ..(((.((.((((....((...((((((((.(((((((..........)))).....).))))))))))..)).((((.....------)))).....)))).))--)))---- ( -36.60, z-score = -0.06, R) >droWil1.scaffold_180699 1893771 110 + 2593675 UUGUGGCUCUCUGUUUAGUUUUCGGCCACGUGCCGCGCAUAUUAAGUGUUAGAAG--GCGCCACCACCGCCGCA-UACCAACCAA-AUGCCGCACAACUCGAACUGAAAACUGA ..............(((((((((((....(((((..((((.....)))).....)--)))).......((.(((-(.........-)))).))..........))))))))))) ( -26.20, z-score = 0.42, R) >droVir3.scaffold_12875 7001741 91 - 20611582 -UGUAGC--UCUGUUUAGUUUUGGUCCACGUGCUUGGCAUAAUAAGCGCUGCCAG--CAGCCG--GCUGCCGAC-UGCCGGCGGC-GUCUCUCGAGAAUU-------------- -.....(--(((((..(((..((.....)).)))..)))......((((((((.(--(((.((--(...))).)-))).))))))-)).....)))....-------------- ( -31.90, z-score = -0.12, R) >consensus UGGUGGUCGUGUGUUUAGUUUUCGGCCACGUGCCCCGCAUAAUAAUAAGCGGCAUUCGAGCCGCUGCAGUUGCAGUUGUAGCUGCGAUGCCACAAUGCCACAAAAGCUGC____ ..(((((((.............)))))))(((....((((.......((((((......))))))((((((((....)))))))).)))))))..................... (-23.18 = -25.20 + 2.02)
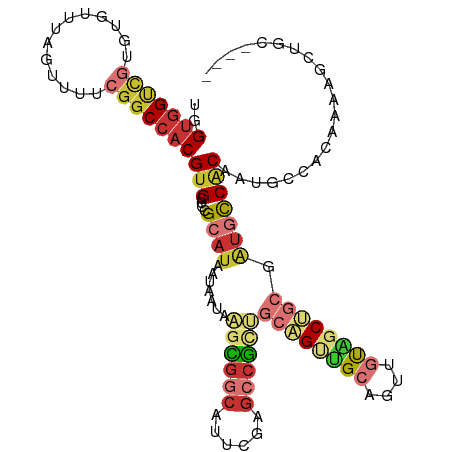

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:32 2011