
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,823,790 – 18,823,898 |
| Length | 108 |
| Max. P | 0.703942 |

| Location | 18,823,790 – 18,823,898 |
|---|---|
| Length | 108 |
| Sequences | 14 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.63 |
| Shannon entropy | 0.40716 |
| G+C content | 0.61809 |
| Mean single sequence MFE | -45.74 |
| Consensus MFE | -29.31 |
| Energy contribution | -29.11 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.703942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18823790 108 - 21146708 UGGCGGCGGCAUGCGUCGCAACCGCGAGAAGUGCUACAAAUGCAACCAAUUUGGGCACUUCGCACGUGCCUGCCCUGAGGAGGCCGAGCGCUGCUACCGCUGCAACGG .(((((..(((.((.((((....))))((((((((.(((((.......)))))))))))))((.((.((((.((....)))))))).)))))))..)))))....... ( -50.30, z-score = -2.21, R) >droSim1.chr2R 17419805 108 - 19596830 UGGCGGCGGCAUGCGUCGCAACCGCGAGAAGUGCUACAAAUGCAACCAAUUCGGGCACUUCGCACGUGCCUGCCCCGAGGAGGCCGAGCGCUGCUACCGCUGCAACGG .(((((..(((.((.((((....))))((((((((...(((.......)))..))))))))((.((.((((.((....)))))))).)))))))..)))))....... ( -47.00, z-score = -1.01, R) >droSec1.super_9 2122221 108 - 3197100 UGGCGGCGGCAUGCGUCGCAACCGCGAGAAGUGCUACAAAUGCAACCAAUUCGGGCACUUCGCACGUGCCUGCCCCGAGGAGGCCGAGCGCUGCUACCGCUGCAACGG .(((((..(((.((.((((....))))((((((((...(((.......)))..))))))))((.((.((((.((....)))))))).)))))))..)))))....... ( -47.00, z-score = -1.01, R) >droYak2.chr2R 18789062 108 - 21139217 UGGCGGCGGCAUGCGUCGCAACCGCGAGAAGUGCUACAAAUGCAACCAGUUUGGGCACUUCGCACGUGCCUGCCCCGAGGAGGCCGAGCGCUGCUACCGUUGCAACGG ((((((((.(.(((.((((....))))((((((((.(((((.......))))))))))))))))((.((((.((....)))))))).)))))))))((((....)))) ( -48.20, z-score = -1.20, R) >droEre2.scaffold_4845 20187040 108 - 22589142 UGGCGGCGGCAUGCGUCGCAACCGCGAGAAGUGCUACAAAUGCAACCAGUUCGGGCACUUCGCACGUGCCUGCCCCGAGGAGGCCGAGCGCUGCUACCGCUGCAACGG .(((((..(((.((.((((....))))((((((((.....((....)).....))))))))((.((.((((.((....)))))))).)))))))..)))))....... ( -47.60, z-score = -0.78, R) >droAna3.scaffold_13266 15165465 108 - 19884421 UGGCGGCGGCAUGCGUCGCAACCGCGAAAAGUGCUACAAAUGCAACCAGUUCGGGCACUUUGCACGUGCCUGCCCCGAGGAGGCCGAGCGCUGCUACCGCUGCAACGG .(((((..(((.((.((((....))))((((((((.....((....)).....))))))))((.((.((((.((....)))))))).)))))))..)))))....... ( -44.40, z-score = -0.14, R) >dp4.chr3 1980771 108 + 19779522 UGGCGGCGGUAUGCGCCGCAACCGCGAGAAGUGCUACAAAUGCAACCAAUUUGGGCACUUUGCACGUGCCUGCCCUGAGGAGGCUGAGCGGUGCUACCGCUGCAACGG ..(((((((((.(((((((....(((.((((((((.(((((.......)))))))))))))...)))((((.((....))))))...))))))))))))))))..... ( -55.90, z-score = -4.07, R) >droPer1.super_73 252173 108 - 267513 UGGCGGCGGUAUGCGCCGCAACCGCGAGAAGUGCUACAAAUGCAACCAAUUUGGGCACUUUGCACGUGCCUGCCCUGAGGAGGCUGAGCGGUGCUACCGCUGCAACGG ..(((((((((.(((((((....(((.((((((((.(((((.......)))))))))))))...)))((((.((....))))))...))))))))))))))))..... ( -55.90, z-score = -4.07, R) >droWil1.scaffold_180699 1869174 108 + 2593675 UGGCGGCGGCAUGCGUCGCAAUCGCGAAAAGUGUUACAAAUGCAAUCAAUUUGGGCACUUUGCACGUGCCUGCCCCGAGGAGGCUGAGCGAUGCUAUCGCUGCAACGG ..(((((((...(((((((....(((.((((((((.(((((.......)))))))))))))...)))((((.((....))))))...)))))))..)))))))..... ( -45.10, z-score = -1.86, R) >droMoj3.scaffold_6496 25658308 108 - 26866924 UGGCGGCGGCAUGCGUCGUAAUCGUGAAAAGUGCUACAAAUGCAAUCAAUUUGGGCACUUUGCACGCGCGUGCCCCGAAGAGGCGGAGCGCUGCUAUCGGUGCAACGG ...(((.((((((((.......((((.((((((((.(((((.......))))))))))))).)))))))))))))))......((..((((((....))))))..)). ( -46.71, z-score = -1.93, R) >droGri2.scaffold_15245 532262 108 - 18325388 CGGUGGCGGCAUGCGCCGCAAUCGUGAAAAGUGCUAUAAGUGCAACCAAUUUGGGCACUUUGCACGCGCCUGCCCGGAGGAGGCGGAGCGCUGCUAUCGCUGCAACGG .(((.(((((....))))).)))((((...((((...((((((..((.....)))))))).))))((((((((((....).))))).)))).....))))........ ( -46.90, z-score = -1.04, R) >anoGam1.chr3R 5098865 99 - 53272125 ------GGACUUUGGCCG---UCGCGAGAAGUGUUACAAGUGUAACCAGAUGGGUCACUUCGCCCGUGACUGCAAGGAGGACCUGGACCGGUGCUACCGUUGCAAUGG ------((.(....)))(---(((((.((((((((((....))))((....))..))))))...))))))(((((((((.(((......))).)).)).))))).... ( -33.20, z-score = -0.32, R) >apiMel3.Group9 5843074 105 - 10282195 ---AGGUGGUUUUGCGCGUGGUCGCGACAAGUGCUACAAGUGUAACCAAUUUGGACACUUUGCACGAGAAUGUAAAGAAGAUCAAGAUCUUUGCUAUCGUUGCCAAGG ---..(((((((((((((....)))).)))).)))))((((((..((.....)))))))).((((((....(((((((.........)))))))..))).)))..... ( -30.40, z-score = -1.09, R) >triCas2.ChLG7 12716584 108 + 17478683 CCGCGGGGGCUUCAACCGCAGCCGCGAGAAGUGCCACAAGUGCAACAAGACGGGGCACUACGCGCGCGACUGCAAGGAGGACUCGGCGCGCUGCUACCGCUGCUACGG .(((.(((.((((....(((((((((.(.((((((....((........))..)))))).).)))).).))))...)))).))).))).((.((....)).))..... ( -41.80, z-score = 0.81, R) >consensus UGGCGGCGGCAUGCGUCGCAACCGCGAGAAGUGCUACAAAUGCAACCAAUUUGGGCACUUCGCACGUGCCUGCCCCGAGGAGGCCGAGCGCUGCUACCGCUGCAACGG ..(((((((.......(((....))).((((((((...(((.......)))..))))))))((((((((((.(.....).))))...))).)))..)))))))..... (-29.31 = -29.11 + -0.20)

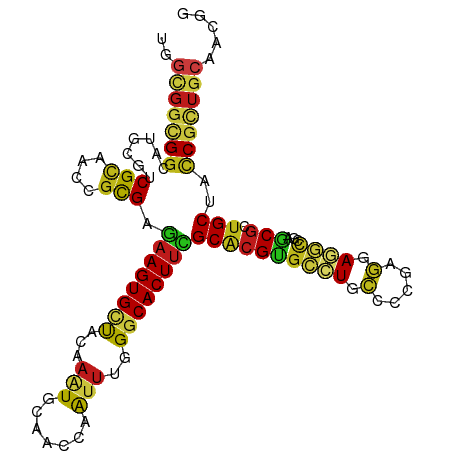

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:31 2011