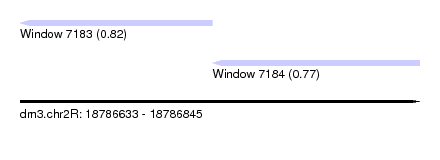
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,786,633 – 18,786,845 |
| Length | 212 |
| Max. P | 0.816896 |

| Location | 18,786,633 – 18,786,735 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 67.09 |
| Shannon entropy | 0.64386 |
| G+C content | 0.46799 |
| Mean single sequence MFE | -29.76 |
| Consensus MFE | -13.16 |
| Energy contribution | -14.21 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.816896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18786633 102 - 21146708 -------CAGCAGAGGCAUUUGU-GUGUCAUUUGGGGCGUGCUGGCCGAGUUUUAUGAGUGCGCGAGCAAUUGGCCAGCAUAAGAAAUUAGCAUUAAAAUCUAAGGUCAG -------...(((((((((....-))))).))))....(((((((((((..........(((....))).)))))))))))..((..((((.((....))))))..)).. ( -33.30, z-score = -2.11, R) >droPer1.super_4 5067397 105 - 7162766 UGGCUUUUGGCUCUCUUUCUAUUAGUAUUACAGGGUAUGUG-UGGACUAGCAAUCGAUCUGUGUCACUCGUUGUCGGGCCAGCUAAGGUCAGAGC-GGACCAGAGAU--- (((((...(((((.....(.((.(((..((((((.....((-(......))).....))))))..))).)).)..)))))))))).((((.....-.))))......--- ( -26.70, z-score = 1.02, R) >droAna3.scaffold_13266 15126282 95 - 19884421 -------UCGCUGAGUAAUUUCUAAUGUCACUUGGGACGCGAUGGUCGAGUGCCC----GCCAAGAUCAACUCGGAGCCACCCGA--UUCGCAUUAAGAUCUAAGGUC-- -------..(((...((..(..((((((.(.(((((..((.....((((((....----..........)))))).))..)))))--.).))))))..)..)).))).-- ( -20.24, z-score = 1.47, R) >droEre2.scaffold_4845 20150925 102 - 22589142 -------CAGCAGAGGCAUUUGU-GUGUCAUUUGGGGCGUGCUGGCCAAGUUUUAUGAGUGCGACAGAAAUUGGCCAGCAUAAGAAAUUAGCAUAAAAAACUAAGGUCAG -------...(((((((((....-))))).))))....(((((((((((.((((...........)))).)))))))))))..((..((((.........))))..)).. ( -28.90, z-score = -1.61, R) >droYak2.chr2R 18751539 96 - 21139217 -------------AGGCAUUUGU-GUGUCACUUGGGGCGUGCUGGCCAAGUUUUAUGAUUGCGAGAGUAAUUGGCCAGCAUAAGAAAUUAGCAUACAAAUCUAAGGUCAG -------------.(((((((((-((((..........((((((((((.........(((((....))))))))))))))).........)))))))))).....))).. ( -32.01, z-score = -2.81, R) >droSec1.super_9 2085229 102 - 3197100 -------CAGCUGAGGCAUUUGU-GUGUCAUUUGGGGCGUGCUGGCCGAGUUUUAUGAGUGCGCGAGCAAUUGGCCAGCAUAAGAAAUUAGCAUUAAAAUCUAAGGUCAG -------..((((((((((....-))))).........(((((((((((..........(((....))).)))))))))))......))))).................. ( -34.40, z-score = -2.13, R) >droSim1.chr2R 17383091 99 - 19596830 -------CAGCAGAGGCAUUUGU-GUGUCAUUUGGGGCGUGCUGGCCAAG-UUUAUA--UGCGCGAGCAAUUGGCCAGCAUAAGAAAUUAGCAUUAAAAUCUAAGGUCAG -------...(((((((((....-))))).))))....(((((((((((.-......--(((....))).)))))))))))..((..((((.((....))))))..)).. ( -32.80, z-score = -2.61, R) >consensus _______CAGCAGAGGCAUUUGU_GUGUCAUUUGGGGCGUGCUGGCCAAGUUUUAUGAGUGCGAGAGCAAUUGGCCAGCAUAAGAAAUUAGCAUUAAAAUCUAAGGUCAG ...............((.........(((......)))(((((((((((..........(((....))).))))))))))).........)).................. (-13.16 = -14.21 + 1.05)



| Location | 18,786,735 – 18,786,845 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 68.01 |
| Shannon entropy | 0.63269 |
| G+C content | 0.36699 |
| Mean single sequence MFE | -20.29 |
| Consensus MFE | -10.69 |
| Energy contribution | -9.54 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.70 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.767512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18786735 110 - 21146708 UUUACAAAAUUUUGCC-CGCAAGUCGUUAACGGCAAAUGC--UUCAAAACUCUCUUCUCCUGCUGAAUCGCGAGAAUAACUUUCGCAAAAAAUCCAUACAAAAAAUUAAUUUU .........((((((.-.(((.((((....))))...)))--............(((((..((......)))))))........))))))....................... ( -16.20, z-score = -1.27, R) >droPer1.super_4 5067502 112 - 7162766 UGUACAGAGUUGUGGCUCAAAGGCCGUUAACGGCAAAUUCCAUUUCUUCAGUUCAUUCAUUUGUUCAUGGCUGGUGGGGGGAUAGCUCAACUUUCAUC-UAACUUUUGGCUUU .....(((((((.((((.....((((....))))....(((.(..(.(((((.(((..........)))))))).)..)))).)))))))))))....-.............. ( -26.10, z-score = -0.27, R) >dp4.chr3 9746959 112 - 19779522 UUUACAGAGUUGUGGCUCAAAGGCCGUUAACGGCAAAUUCCAUUUCUUCAUUUCAUUCAUUUGUUCAUGGCUGGUGGGGGGAUAGCUCAACUUUCAUC-AAACUUUUGGCUUU .....(((((((.((((.....((((....)))).......((..((((((..((..(((......)))..))))))))..)))))))))))))....-.............. ( -26.00, z-score = -0.90, R) >droAna3.scaffold_13266 15126377 93 - 19884421 UUUACAAAAUGUUGCCAAAAAAAUUGUUAACGGCAAAUGC--UUU---------UCUUUUUGCUUUUUUGGGAGGAUAACUUUCCC---AUGGCAAGCCCCGAAAUU------ ...........((((((..............((((((...--...---------....))))))....(((((((.....))))))---)))))))...........------ ( -19.60, z-score = -0.40, R) >droEre2.scaffold_4845 20151027 107 - 22589142 UUUACAAAA-UGUGCC-CGCCAGUCGUUAACGGCAAAUGC--UUUUUAACUCUCUUUUCUUGGUGAAUCGCGAGAAUAACUUUCGC-AAAAAUCCAUA-AAAACAUUGGUUUU .........-......-.((((((.(((((.(((....))--)..)))))....((((..(((......((((((.....))))))-......)))..-)))).))))))... ( -19.90, z-score = -0.81, R) >droYak2.chr2R 18751635 102 - 21139217 UUUACAAAA-UUUGCC-CGCAAGUCGUUAACGGCAAAUGC--UUUUAAACUCUCUUUUCUUGGUGAAUCGCGAGAAUAACUUUCGC-AAAAAUCCAUA-AAAACAUUG----- ........(-((((((-(....)..(....))))))))..--............((((..(((......((((((.....))))))-......)))..-)))).....----- ( -17.50, z-score = -1.15, R) >droSec1.super_9 2085331 109 - 3197100 UUUACAAAA-UUUGCC-CGCAAGUCGUUAACGGCAAAUGC--UUCAAAACUCUCUUUUCUUGCUGAAUCGCGAGAAUAACUUUCGCAAAAAAUCGAUACGAAAAAUUAAUUUU .........-(((((.-.(((.((((....))))...)))--..............(((((((......)))))))........)))))........................ ( -18.50, z-score = -0.80, R) >droSim1.chr2R 17383190 109 - 19596830 UUUACAAAA-UUUGCC-CGCAAGUCGUUAACGGCAAAUGC--UUCAAAACUCUCUUUUCUUGCUGAAUCGCGAGAAUAACUUUCGCAAAAAAUCGAUACGAAAAAUUAAUUUU .........-(((((.-.(((.((((....))))...)))--..............(((((((......)))))))........)))))........................ ( -18.50, z-score = -0.80, R) >consensus UUUACAAAA_UUUGCC_CGCAAGUCGUUAACGGCAAAUGC__UUUAAAACUCUCUUUUCUUGCUGAAUCGCGAGAAUAACUUUCGC_AAAAAUCCAUA_AAAAAAUUGAUUUU ......................((((....))))...................................((((((.....))))))........................... (-10.69 = -9.54 + -1.16)

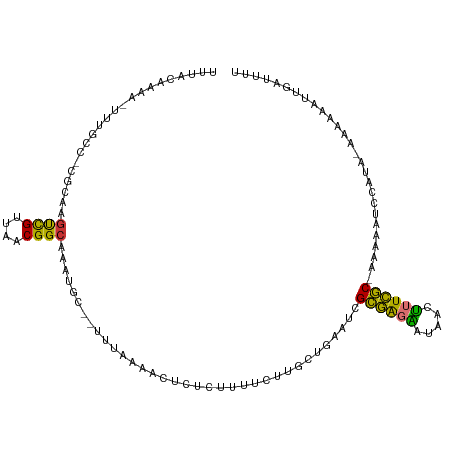
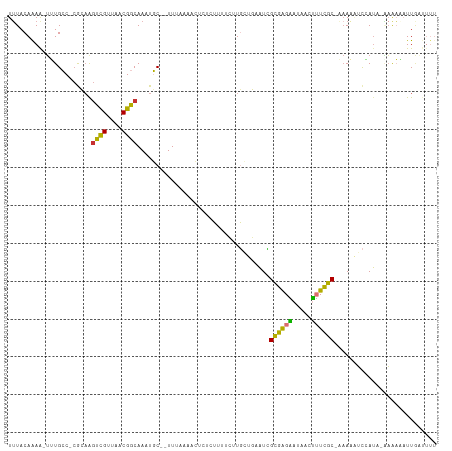
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:27 2011