
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,785,792 – 18,785,883 |
| Length | 91 |
| Max. P | 0.589474 |

| Location | 18,785,792 – 18,785,883 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 79.54 |
| Shannon entropy | 0.37760 |
| G+C content | 0.49903 |
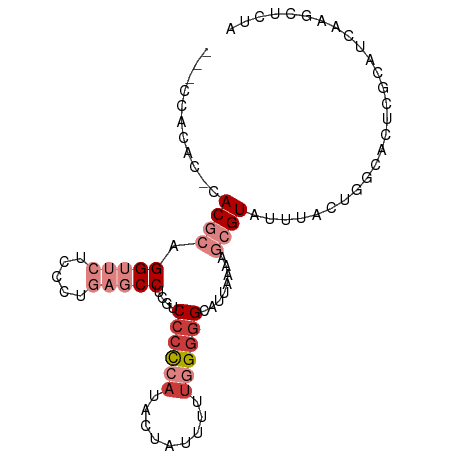
| Mean single sequence MFE | -25.36 |
| Consensus MFE | -12.42 |
| Energy contribution | -14.44 |
| Covariance contribution | 2.02 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
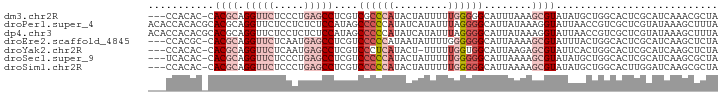
>dm3.chr2R 18785792 91 + 21146708 ---CCACAC-CACGCAGGUUCUCCCUGAGCCUCGUCGCCCAUACUAUUUUUGGGGGCAUUUAAAGCGUAUAUGCUGGCACUCGCAUCAAACGCUA ---......-.(((.((((((.....))))))))).((((...(.......).))))......(((((..((((........))))...))))). ( -27.30, z-score = -1.93, R) >droPer1.super_4 5066448 95 + 7162766 ACACCACACGCACGCAGGUUCUCCUCUCUCCAUAGCCCCCAUAUCAUAUUUAGGGGCAUUAUAAAGGUAUUAACCGUCGCUCGUAUAAAGCUUUA .......(((..((..((((..(((.........(((((.............))))).......)))....))))..))..)))........... ( -16.91, z-score = -0.70, R) >dp4.chr3 9745940 95 + 19779522 ACACCACACGCACGCAGGUUCUCCUCUCUCCAUAGCCCCCAUAUCAUAUUUAGGGGCAUUAUAAAGGUAUUAACCGUCGCUCGUAUAAAGCUUUA .......(((..((..((((..(((.........(((((.............))))).......)))....))))..))..)))........... ( -16.91, z-score = -0.70, R) >droEre2.scaffold_4845 20150091 91 + 22589142 ---CCACGC-CACGCAGGUUCUCAAUGAGCCUCGUCCCCCAUAAUAUUUUGGGGGGCAUUAAAAGCGUAUUUACUGGCACUCGCAUCAAGCUCUA ---....((-(((((((((((.....)))))).((((((((........)))))))).......)))).......)))....((.....)).... ( -33.71, z-score = -4.32, R) >droYak2.chr2R 18750689 90 + 21139217 ---CCACAC-CACGCAGGUUCUCAAUGAGCCUCGUCCCUCAUACU-UUUUUGGUGGCAUUAAGAGCGUAUUCACUGGCACUCGCAUCAAGCUCUA ---((((..-.(((.((((((.....)))))))))....((....-....)))))).....(((((((....))..((....)).....))))). ( -21.70, z-score = -1.38, R) >droSec1.super_9 2084377 91 + 3197100 ---UCACAC-CACGCAGGUUCUCCCUGAGCCUCGUCCCCCAUACUAUUUUUGGGGGCAUUAAAAGCGUAUAUGCUGGCACUCGCAUCAAGCGCUA ---......-.(((.((((((.....)))))))))((((((.........)))))).......(((((..((((........))))...))))). ( -30.50, z-score = -2.90, R) >droSim1.chr2R 17382258 91 + 19596830 ---CCACAC-CACGCAGGUUCUCCCUGAGCCUCGUCCCCCAUACUAUUUUUGGGGGCAUUAAAAGCGUAUAUGCUGGCACUUGGAUCAAGCGCUA ---......-.((((((((((.....)))))).(.((((((.........))))))).......))))......((((.((((...)))).)))) ( -30.50, z-score = -2.19, R) >consensus ___CCACAC_CACGCAGGUUCUCCCUGAGCCUCGUCCCCCAUACUAUUUUUGGGGGCAUUAAAAGCGUAUUUACUGGCACUCGCAUCAAGCUCUA ...........((((.(((((.....)))))....((((((.........))))))........))))........................... (-12.42 = -14.44 + 2.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:25 2011