| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,755,064 – 18,755,202 |
| Length | 138 |
| Max. P | 0.939093 |

| Location | 18,755,064 – 18,755,202 |
|---|---|
| Length | 138 |
| Sequences | 10 |
| Columns | 150 |
| Reading direction | reverse |
| Mean pairwise identity | 73.31 |
| Shannon entropy | 0.56590 |
| G+C content | 0.48778 |
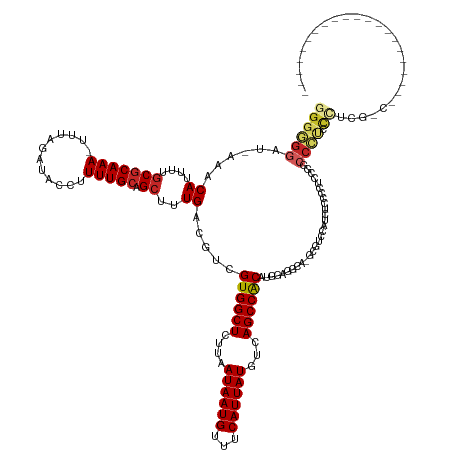
| Mean single sequence MFE | -36.75 |
| Consensus MFE | -19.11 |
| Energy contribution | -19.31 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.939093 |
| Prediction | RNA |
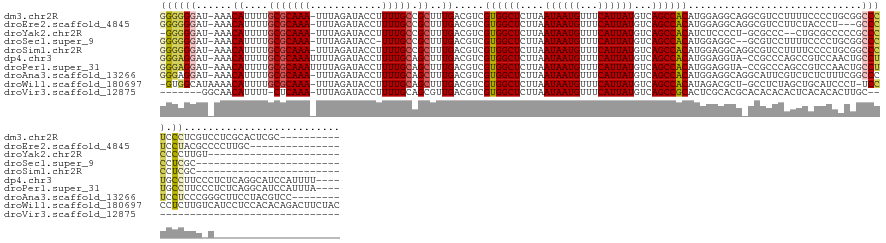
Download alignment: ClustalW | MAF
>dm3.chr2R 18755064 138 - 21146708 GGGGGGAU-AAACAUUUUGCGCAAA-UUUAGAUACCUUUUGCCGCUUUGACGUCGUGGCUCUUAAUAAUGUUUCAUUAUGUCAGCCACAUGGAGGCAGGCGUCCUUUUCCCCUGCGGCCCUCCCUCGUCCUCGCACUCGC---------- (.((((((-...((....(((((((-...........)))).)))..)).(((.((((((....((((((...))))))...)))))))))((((.(((.((((.........).)))))).)))))))))).)......---------- ( -41.00, z-score = -0.81, R) >droEre2.scaffold_4845 20119402 130 - 22589142 GGGGGGAU-AAACAUUUUGCGCAAA-UUUAGAUACCUUUUGCCGCUUUGACGUCGUGGCUCUUAAUAAUGUUUCAUUAUGUCAGCCACAUGGAGGCAGGCGUCCUUCUACCCU---GCCCUCCUACGCCCCUUGC--------------- ((((((..-...((....(((((((-...........)))).)))..)).(((.((((((....((((((...))))))...))))))..(((((((((.((......)))))---).))))).)))))))))..--------------- ( -43.90, z-score = -2.84, R) >droYak2.chr2R 18719893 122 - 21139217 -GGGGGAU-AAACAUUUUGCGCAAA-UUUAGAUACCUUUUGCCGCUUUGACGUCGUGGCUCUUAAUAAUGUUUCAUUAUGUCAGCCACAUCUCCCCU-GCGCCC--CUGCGCCCCCGCCCCCCCUUGU---------------------- -(((((..-.........(((((..-...((((.......(.((......)).)((((((....((((((...))))))...))))))))))....)-))))..--..(((....))).)))))....---------------------- ( -34.50, z-score = -2.76, R) >droSec1.super_9 2053824 121 - 3197100 GGGGGGAU-AAACAUUUUGCGCAAA-UUUAGAUACC-UUUGCCGCUUUGACGUCGUGGCUCUUAAUAAUGUUUCAUUAUGUCAGCCACAUGGAGGC--GCGUCCUUUUCCCCUGCGGCCCCCUCGC------------------------ ((((((..-......((((......-..))))....-...(((((.....(((.((((((....((((((...))))))...)))))))))((((.--....)))).......)))))))))))..------------------------ ( -35.10, z-score = -0.36, R) >droSim1.chr2R 17350251 124 - 19596830 GGGGGGAU-AAACAUUUUGCGCAAA-UUUAGAUACCUUUUGCCGCUUUGACGUCGUGGCUCUUAAUAAUGUUUCAUUAUGUCAGCCACAUGGAGGCAGGCGUCCUUUUCCCCUGCGGCCCCCUCGC------------------------ ((((((..-...((....(((((((-...........)))).)))..))..(((((((((....((((((...))))))...))))))......(((((.(.......).))))))))))))))..------------------------ ( -40.30, z-score = -1.49, R) >dp4.chr3 17454034 144 - 19779522 GGGAGGAU-AAACAUUUUGCGCAAAUUUUAGAUACCUUUUGCAGCUUUGACGUCGUGGCUCUUAAUAAUGUUUCAUUAUGUCAGCCACAUGGAGGUA-CCGCCCAGCCGUCCAACUGCCUUGCCUUCCCUCUCAGGCAUCCAUUUU---- ((.(((((-...((....(((((((............))))).))..))..)))((((((....((((((...))))))...)))))).(((((((.-.......))).)))).)).)).(((((........)))))........---- ( -36.40, z-score = -0.65, R) >droPer1.super_31 597542 144 + 935084 GGGAGGAU-AAACAUUUUGCGCAAAUUUUAGAUACCUUUUGCAGCUUUGACGUCGUGGCUCUUAAUAAUGUUUCAUUAUGUCAGCCACAUGGAGGUA-CCGCCCAGCCGUCCAACUGCCUUGCCUUCCCUCUCAGGCAUCCAUUUA---- ((.(((((-...((....(((((((............))))).))..))..)))((((((....((((((...))))))...)))))).(((((((.-.......))).)))).)).)).(((((........)))))........---- ( -36.40, z-score = -0.63, R) >droAna3.scaffold_13266 15092443 140 - 19884421 GGGAGGAU-AAACAUUUUGCGCAAA-UUUAGAUACCUUUUGCAGCUUUGACGUCGUGGCUCUUAAUAAUGUUUCAUUAUGUCAGCCACAUGGAGGCAGGCAUUCGUCUCUCUUUCGGCCCUCCUCCCGGGCUUCCUACGUCC-------- (((((((.-...((....(((((((-...........))))).))..))..(((((((((....((((((...))))))...))))))..((((((........)))))).....)))..)))))))((((.......))))-------- ( -44.10, z-score = -2.43, R) >droWil1.scaffold_180697 3551220 146 + 4168966 -GUGGCAUAAAACAUUUUGCGCAAA-UUUAGAUACCUUUUGCAGCUUUGACGUCGUGGCUCUUAAUAAUGUUUCAUUAUGUCAGCCACAUAGACGCU-GCCUCUAGCUGCAUCCCU-UCCCCUCUUGUCAUCCUCCACACAGACUUCUAC -((((.............(((((((-...........))))).))..(((((((((((((....((((((...))))))...))))))...)))((.-((.....)).))......-.........))))....))))............ ( -32.40, z-score = -2.17, R) >droVir3.scaffold_12875 19164182 109 - 20611582 -------GGCAACAUUUU-CUCAAA-UUUAGAUACCUUUUGCAGCGUUGACGUCGUGGCUCUUAAUAAUGUUUCAUUAUGUCAGCCGCACUCGCACGCACACACACUCACACACUUGC-------------------------------- -------.((((.....(-((....-...)))......)))).(((((((.((.((((((....((((((...))))))...))))))))))).))))....................-------------------------------- ( -23.40, z-score = -1.54, R) >consensus GGGGGGAU_AAACAUUUUGCGCAAA_UUUAGAUACCUUUUGCAGCUUUGACGUCGUGGCUCUUAAUAAUGUUUCAUUAUGUCAGCCACAUGGAGGCA_GCGUCCAUCUCCCCUCCGGCCCUCCCUCG_C_____________________ ..((((......((....(((((((............))))).))..)).....((((((....((((((...))))))...)))))).............................))))............................. (-19.11 = -19.31 + 0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:22 2011